Genetic Plurality of OXA/NDM-Encoding Features Characterized From Enterobacterales Recovered From Czech Hospitals
- PMID: 33633720
- PMCID: PMC7900173
- DOI: 10.3389/fmicb.2021.641415
Genetic Plurality of OXA/NDM-Encoding Features Characterized From Enterobacterales Recovered From Czech Hospitals
Abstract
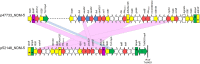
The aim of this study was to characterize four Enterobacterales co-producing NDM- and OXA-48-like carbapenemases from Czech patients with travel history or/and previous hospitalization abroad. Klebsiella pneumoniae isolates belonged to "high risk" clones ST147, ST11, and ST15, while the Escherichia coli isolate was assigned to ST167. All isolates expressed resistance against most β-lactams, including carbapenems, while retaining susceptibility to colistin. Furthermore, analysis of WGS data showed that all four isolates co-produced OXA-48- and NDM-type carbapenemases, in different combinations (Kpn47733: bla NDM- 5 + bla OXA- 181; Kpn50595: bla NDM- 1 + bla OXA- 181; Kpn51015: bla NDM- 1 + bla OXA- 244; Eco52418: bla NDM- 5 + bla OXA- 244). In Kpn51015, the bla OXA- 244 was found on plasmid p51015_OXA-244, while the respective gene was localized in the chromosomal contig of E. coli Eco52418. On the other hand, bla OXA- 181 was identified on a ColKP3 plasmid in isolate Kpn47733, while a bla OXA- 181-carrying plasmid being an IncX3-ColKP3 fusion was identified in Kpn50595. The bla NDM- 1 gene was found on two different plasmids, p51015_NDM-1 belonging to a novel IncH plasmid group and p51015_NDM-1 being an IncF K 1-FIB replicon. Furthermore, the bla NDM- 5 was found in two IncFII plasmids exhibiting limited nucleotide similarity to each other. In both plasmids, the genetic environment of bla NDM- 5 was identical. Finally, in all four carbapenemase-producing isolates, a diverse number of additional replicons, some of these associated with important resistance determinants, like bla CTX-M- 15, arr-2 and ermB, were identified. In conclusion, this study reports the first description of OXA-244-producing Enterobacterales isolated from Czech hospitals. Additionally, our findings indicated the genetic plurality involved in the acquisition and dissemination of determinants encoding OXA/NDM carbapenemases.
Keywords: NDM-1; NDM-5; OXA-181; OXA-244; mobile genetic elements.
Copyright © 2021 Chudejova, Kraftova, Mattioni Marchetti, Hrabak, Papagiannitsis and Bitar.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Ahmad N., Ali S. M., Khan A. U. (2019). Molecular characterization of novel sequence type of carbapenem-resistant New Delhi metallo-β-lactamase-1-producing Klebsiella pneumoniae in the neonatal intensive care unit of an Indian hospital. Int. J. Antimicrob. Agents 53 525–529. 10.1016/j.ijantimicag.2018.12.005 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

