Transcriptomic Analysis of Rat Macrophages
- PMID: 33633725
- PMCID: PMC7902030
- DOI: 10.3389/fimmu.2020.594594
Transcriptomic Analysis of Rat Macrophages
Abstract
The laboratory rat is widely used as a model for human diseases. Many of these diseases involve monocytes and tissue macrophages in different states of activation. Whilst methods for in vitro differentiation of mouse macrophages from embryonic stem cells (ESC) and bone marrow (BM) are well established, these are lacking for the rat. The gene expression profiles of rat macrophages have also not been characterised to the same extent as mouse. We have established the methodology for production of rat ESC-derived macrophages and compared their gene expression profiles to macrophages obtained from the lung and peritoneal cavity and those differentiated from BM and blood monocytes. We determined the gene signature of Kupffer cells in the liver using rats deficient in macrophage colony stimulating factor receptor (CSF1R). We also examined the response of BM-derived macrophages to lipopolysaccharide (LPS). The results indicate that many, but not all, tissue-specific adaptations observed in mice are conserved in the rat. Importantly, we show that unlike mice, rat macrophages express the CSF1R ligand, colony stimulating factor 1 (CSF1).
Keywords: Kupffer cell; colony-stimulating factor 1 receptor; lipopolysaccharide; macrophage; rat.
Copyright © 2021 Pridans, Irvine, Davis, Lefevre, Bush and Hume.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
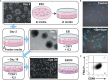
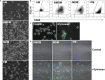




References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

