Human Ubiquitin-Specific Peptidase 18 Is Regulated by microRNAs via the 3'Untranslated Region, A Sequence Duplicated in Long Intergenic Non-coding RNA Genes Residing in chr22q11.21
- PMID: 33633774
- PMCID: PMC7901961
- DOI: 10.3389/fgene.2020.627007
Human Ubiquitin-Specific Peptidase 18 Is Regulated by microRNAs via the 3'Untranslated Region, A Sequence Duplicated in Long Intergenic Non-coding RNA Genes Residing in chr22q11.21
Abstract
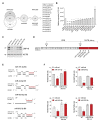
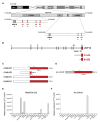
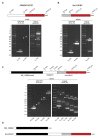
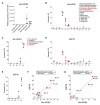
Ubiquitin-specific peptidase 18 (USP18) acts as gatekeeper of type I interferon (IFN) responses by binding to the IFN receptor subunit IFNAR2 and preventing activation of the downstream JAK/STAT pathway. In any given cell type, the level of USP18 is a key determinant of the output of IFN-stimulated transcripts. How the baseline level of USP18 is finely tuned in different cell types remains ill defined. Here, we identified microRNAs (miRNAs) that efficiently target USP18 through binding to the 3'untranslated region (3'UTR). Among these, three miRNAs are particularly enriched in circulating monocytes which exhibit low baseline USP18. Intriguingly, the USP18 3'UTR sequence is duplicated in human and chimpanzee genomes. In humans, four USP18 3'UTR copies were previously found to be embedded in long intergenic non-coding (linc) RNA genes residing in chr22q11.21 and known as FAM247A-D. Here, we further characterized their sequence and measured their expression profile in human tissues. Importantly, we describe an additional lincRNA bearing USP18 3'UTR (here linc-UR-B1) that is expressed only in testis. RNA-seq data analyses from testicular cell subsets revealed a positive correlation between linc-UR-B1 and USP18 expression in spermatocytes and spermatids. Overall, our findings uncover a set of miRNAs and lincRNAs, which may be part of a network evolved to fine-tune baseline USP18, particularly in cell types where IFN responsiveness needs to be tightly controlled.
Keywords: 22q11.2; 3'untranslated region, microRNAs; Testis; long intergenic non-coding RNA; type I interferon; ubiquitin-specific peptidase.
Copyright © 2021 Rubino, Cruciani, Tchitchek, Le Tortorec, Rolland, Veli, Vallet, Gaggi, Michel, Dejucq-Rainsford and Pellegrini.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Insulin receptor substrate-4 interacts with ubiquitin-specific protease 18 to activate the Jak/STAT signaling pathway.Oncotarget. 2017 Nov 18;8(62):105923-105935. doi: 10.18632/oncotarget.22510. eCollection 2017 Dec 1. Oncotarget. 2017. PMID: 29285303 Free PMC article.
-
Birth of a Regulatory Long Non-coding RNA/Gene, linc-UR-UB.Front Genet. 2021 Apr 30;12:661425. doi: 10.3389/fgene.2021.661425. eCollection 2021. Front Genet. 2021. PMID: 33995491 Free PMC article.
-
Negative regulation of the RLR-mediated IFN signaling pathway by duck ubiquitin-specific protease 18 (USP18).J Cell Physiol. 2019 Apr;234(4):3995-4004. doi: 10.1002/jcp.27208. Epub 2018 Sep 7. J Cell Physiol. 2019. PMID: 30256391
-
Emerging Roles of USP18: From Biology to Pathophysiology.Int J Mol Sci. 2020 Sep 17;21(18):6825. doi: 10.3390/ijms21186825. Int J Mol Sci. 2020. PMID: 32957626 Free PMC article. Review.
-
Functions of long intergenic non-coding (linc) RNAs in plants.J Plant Res. 2017 Jan;130(1):67-73. doi: 10.1007/s10265-016-0894-0. Epub 2016 Dec 20. J Plant Res. 2017. PMID: 27999969 Review.
Cited by
-
The Effects of Combined Exposure to Bisphenols and Perfluoroalkyls on Human Perinatal Stem Cells and the Potential Implications for Health Outcomes.Int J Mol Sci. 2023 Oct 9;24(19):15018. doi: 10.3390/ijms241915018. Int J Mol Sci. 2023. PMID: 37834465 Free PMC article.
-
IFN-I inducible miR-3614-5p targets ADAR1 isoforms and fine tunes innate immune activation.Front Immunol. 2022 Jul 22;13:939907. doi: 10.3389/fimmu.2022.939907. eCollection 2022. Front Immunol. 2022. PMID: 35935998 Free PMC article.
-
An ancestral genomic sequence that serves as a nucleation site for de novo gene birth.PLoS One. 2022 May 12;17(5):e0267864. doi: 10.1371/journal.pone.0267864. eCollection 2022. PLoS One. 2022. PMID: 35552551 Free PMC article.
-
The Non-Coding RNA Journal Club: Highlights on Recent Papers-10.Noncoding RNA. 2022 Jan 10;8(1):3. doi: 10.3390/ncrna8010003. Noncoding RNA. 2022. PMID: 35076559 Free PMC article.
-
Human mesenchymal amniotic fluid stem cells reveal an unexpected neuronal potential differentiating into functional spinal motor neurons.Front Cell Dev Biol. 2022 Jul 22;10:936990. doi: 10.3389/fcell.2022.936990. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35938174 Free PMC article.
References
-
- Allantaz F., Cheng D. T., Bergauer T., Ravindran P., Rossier M. F., Ebeling M., et al. . (2012). Expression profiling of human immune cell subsets identifies miRNA-mRNA regulatory relationships correlated with cell type specific expression. PLoS One 7:e29979. 10.1371/journal.pone.0029979, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

