Common genetic signatures of Alzheimer's disease in Down Syndrome
- PMID: 33633844
- PMCID: PMC7871416
- DOI: 10.12688/f1000research.27096.2
Common genetic signatures of Alzheimer's disease in Down Syndrome
Abstract
Background: People with Down Syndrome (DS) are born with an extra copy of Chromosome (Chr) 21 and many of these individuals develop Alzheimer's Disease (AD) when they age. This is due at least in part to the extra copy of the APP gene located on Chr 21. By 40 years, most people with DS have amyloid plaques which disrupt brain cell function and increase their risk for AD. About half of the people with DS develop AD and the associated dementia around 50 to 60 years of age, which is about the age at which the hereditary form of AD, early onset AD, manifests. In the absence of Chr 21 trisomy, duplication of APP alone is a cause of early onset Alzheimer's disease, making it likely that having three copies of APP is important in the development of AD and in DS. Methods: We investigate the relationship between AD and DS through integrative analysis of genesets derived from a MeSH query of AD and DS associated beta amyloid peptides, Chr 21, GWAS identified AD risk factor genes, and differentially expressed genes in individuals with DS. Results: Unique and shared aspects of each geneset were evaluated based on functional enrichment analysis, transcription factor profile and network interactions. Genes that may be important to both disorders in the context of direct association with APP processing, Tau post translational modification and network connectivity are ACSM1, APBA2, APLP1, BACE2, BCL2L, COL18A1, DYRK1A, IK, KLK6, METTL2B, MTOR, NFE2L2, NFKB1, PRSS1, QTRT1, RCAN1, RUNX1, SAP18 SOD1, SYNJ1, S100B. Conclusions: Our findings confirm that oxidative stress, apoptosis, inflammation and immune system processes likely contribute to the pathogenesis of AD and DS which is consistent with other published reports.
Keywords: Alzheimer’s Disease; Behavior; Down Syndrome; Learning; Memory.
Copyright: © 2021 Sharma A et al.
Conflict of interest statement
No competing interests were disclosed.
Figures

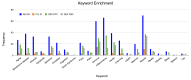
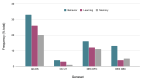
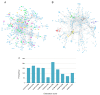
Similar articles
-
Genetic Mapping of APP and Amyloid-β Biology Modulation by Trisomy 21.J Neurosci. 2022 Aug 17;42(33):6453-6468. doi: 10.1523/JNEUROSCI.0521-22.2022. Epub 2022 Jul 14. J Neurosci. 2022. PMID: 35835549 Free PMC article.
-
BACE2, as a novel APP theta-secretase, is not responsible for the pathogenesis of Alzheimer's disease in Down syndrome.FASEB J. 2006 Jul;20(9):1369-76. doi: 10.1096/fj.05-5632com. FASEB J. 2006. PMID: 16816112
-
[Elucidating Pathogenic Mechanisms of Early-onset Alzheimer's Disease in Down Syndrome Patients].Yakugaku Zasshi. 2017;137(7):801-805. doi: 10.1248/yakushi.16-00236-2. Yakugaku Zasshi. 2017. PMID: 28674290 Review. Japanese.
-
Trisomy of human chromosome 21 enhances amyloid-β deposition independently of an extra copy of APP.Brain. 2018 Aug 1;141(8):2457-2474. doi: 10.1093/brain/awy159. Brain. 2018. PMID: 29945247 Free PMC article.
-
Untangle the mystery behind DS-associated AD - Is APP the main protagonist?Ageing Res Rev. 2023 Jun;87:101930. doi: 10.1016/j.arr.2023.101930. Epub 2023 Apr 7. Ageing Res Rev. 2023. PMID: 37031726 Review.
Cited by
-
Causal Discovery with Generalized Linear Models through Peeling Algorithms.J Mach Learn Res. 2024;25:310. J Mach Learn Res. 2024. PMID: 39758585 Free PMC article.
-
Triplication of Synaptojanin 1 in Alzheimer's Disease Pathology in Down Syndrome.Curr Alzheimer Res. 2022;19(12):795-807. doi: 10.2174/1567205020666221202102832. Curr Alzheimer Res. 2022. PMID: 36464875
-
Gene targeting in amyotrophic lateral sclerosis using causality-based feature selection and machine learning.Mol Med. 2023 Jan 24;29(1):12. doi: 10.1186/s10020-023-00603-y. Mol Med. 2023. PMID: 36694130 Free PMC article.
-
Dynamic Changes in Central and Peripheral Neuro-Injury vs. Neuroprotective Serum Markers in COVID-19 Are Modulated by Different Types of Anti-Viral Treatments but Do Not Affect the Incidence of Late and Early Strokes.Biomedicines. 2021 Nov 29;9(12):1791. doi: 10.3390/biomedicines9121791. Biomedicines. 2021. PMID: 34944606 Free PMC article.
-
Dual-Specificity, Tyrosine Phosphorylation-Regulated Kinases (DYRKs) and cdc2-Like Kinases (CLKs) in Human Disease, an Overview.Int J Mol Sci. 2021 Jun 3;22(11):6047. doi: 10.3390/ijms22116047. Int J Mol Sci. 2021. PMID: 34205123 Free PMC article. Review.
References
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

