MetaHiC phage-bacteria infection network reveals active cycling phages of the healthy human gut
- PMID: 33634788
- PMCID: PMC7963479
- DOI: 10.7554/eLife.60608
MetaHiC phage-bacteria infection network reveals active cycling phages of the healthy human gut
Abstract
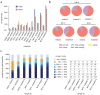
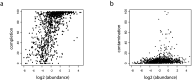
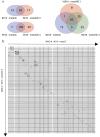
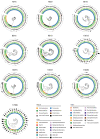
Bacteriophages play important roles in regulating the intestinal human microbiota composition, dynamics, and homeostasis, and characterizing their bacterial hosts is needed to understand their impact. We applied a metagenomic Hi-C approach on 10 healthy human gut samples to unveil a large infection network encompassing more than 6000 interactions bridging a metagenomic assembled genomes (MAGs) and a phage sequence, allowing to study in situ phage-host ratio. Whereas three-quarters of these sequences likely correspond to dormant prophages, 5% exhibit a much higher coverage than their associated MAG, representing potentially actively replicating phages. We detected 17 sequences of members of the crAss-like phage family, whose hosts diversity remained until recently relatively elusive. For each of them, a unique bacterial host was identified, all belonging to different genus of Bacteroidetes. Therefore, metaHiC deciphers infection network of microbial population with a high specificity paving the way to dynamic analysis of mobile genetic elements in complex ecosystems.
Keywords: Hi-C; bacteroidetes; computational biology; human; infectious disease; metagenomics; microbiology; microbiome; proximity ligation; virus.
© 2021, Marbouty et al.
Conflict of interest statement
MM, AT, GM, RK No competing interests declared
Figures



Similar articles
-
Novel canine high-quality metagenome-assembled genomes, prophages and host-associated plasmids provided by long-read metagenomics together with Hi-C proximity ligation.Microb Genom. 2022 Mar;8(3):000802. doi: 10.1099/mgen.0.000802. Microb Genom. 2022. PMID: 35298370 Free PMC article.
-
Prophages in Lactobacillus reuteri Are Associated with Fitness Trade-Offs but Can Increase Competitiveness in the Gut Ecosystem.Appl Environ Microbiol. 2019 Dec 13;86(1):e01922-19. doi: 10.1128/AEM.01922-19. Print 2019 Dec 13. Appl Environ Microbiol. 2019. PMID: 31676478 Free PMC article.
-
Discovery, diversity, and functional associations of crAss-like phages in human gut metagenomes from four Dutch cohorts.Cell Rep. 2022 Jan 11;38(2):110204. doi: 10.1016/j.celrep.2021.110204. Cell Rep. 2022. PMID: 35021085
-
The Human Gut Phage Community and Its Implications for Health and Disease.Viruses. 2017 Jun 8;9(6):141. doi: 10.3390/v9060141. Viruses. 2017. PMID: 28594392 Free PMC article. Review.
-
Phage hunters: Computational strategies for finding phages in large-scale 'omics datasets.Virus Res. 2018 Jan 15;244:110-115. doi: 10.1016/j.virusres.2017.10.019. Epub 2017 Nov 1. Virus Res. 2018. PMID: 29100906 Review.
Cited by
-
Sequence analysis of Discoknowium, an A5 mycobacteriophage.Microbiol Resour Announc. 2024 Jan 17;13(1):e0092023. doi: 10.1128/MRA.00920-23. Epub 2023 Dec 4. Microbiol Resour Announc. 2024. PMID: 38047653 Free PMC article.
-
ViralCC retrieves complete viral genomes and virus-host pairs from metagenomic Hi-C data.Nat Commun. 2023 Jan 31;14(1):502. doi: 10.1038/s41467-023-35945-y. Nat Commun. 2023. PMID: 36720887 Free PMC article.
-
Hi-C metagenome sequencing reveals soil phage-host interactions.Nat Commun. 2023 Nov 23;14(1):7666. doi: 10.1038/s41467-023-42967-z. Nat Commun. 2023. PMID: 37996432 Free PMC article.
-
Phage-microbe dynamics after sterile faecal filtrate transplantation in individuals with metabolic syndrome: a double-blind, randomised, placebo-controlled clinical trial assessing efficacy and safety.Nat Commun. 2023 Sep 12;14(1):5600. doi: 10.1038/s41467-023-41329-z. Nat Commun. 2023. PMID: 37699894 Free PMC article. Clinical Trial.
-
Interactions of Bacteriophages with Animal and Human Organisms-Safety Issues in the Light of Phage Therapy.Int J Mol Sci. 2021 Aug 19;22(16):8937. doi: 10.3390/ijms22168937. Int J Mol Sci. 2021. PMID: 34445641 Free PMC article. Review.
References
-
- Andrews S. 0.11.9FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010 http://www.bioinformatics.babraham.ac.uk/projects/fastqc
-
- Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, Bertalan M, Borruel N, Casellas F, Fernandez L, Gautier L, Hansen T, Hattori M, Hayashi T, Kleerebezem M, Kurokawa K, Leclerc M, Levenez F, Manichanh C, Nielsen HB, Nielsen T, Pons N, Poulain J, Qin J, Sicheritz-Ponten T, Tims S, Torrents D, Ugarte E, Zoetendal EG, Wang J, Guarner F, Pedersen O, de Vos WM, Brunak S, Doré J, Antolín M, Artiguenave F, Blottiere HM, Almeida M, Brechot C, Cara C, Chervaux C, Cultrone A, Delorme C, Denariaz G, Dervyn R, Foerstner KU, Friss C, van de Guchte M, Guedon E, Haimet F, Huber W, van Hylckama-Vlieg J, Jamet A, Juste C, Kaci G, Knol J, Lakhdari O, Layec S, Le Roux K, Maguin E, Mérieux A, Melo Minardi R, M'rini C, Muller J, Oozeer R, Parkhill J, Renault P, Rescigno M, Sanchez N, Sunagawa S, Torrejon A, Turner K, Vandemeulebrouck G, Varela E, Winogradsky Y, Zeller G, Weissenbach J, Ehrlich SD, Bork P, MetaHIT Consortium Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. - DOI - PMC - PubMed
-
- Blondel VD, Guillaume J-L, Lambiotte R, Lefebvre E. Fast unfolding of communities in large networks. Journal of Statistical Mechanics: Theory and Experiment. 2008;2008:P10008. doi: 10.1088/1742-5468/2008/10/P10008. - DOI
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

