Dissecting the role of TP53 alterations in del(11q) chronic lymphocytic leukemia
- PMID: 33634999
- PMCID: PMC7862176
- DOI: 10.1002/ctm2.304
Dissecting the role of TP53 alterations in del(11q) chronic lymphocytic leukemia
Abstract
Background: Several genetic alterations have been identified as driver events in chronic lymphocytic leukemia (CLL) pathogenesis and oncogenic evolution. Concurrent driver alterations usually coexist within the same tumoral clone, but how the cooperation of multiple genomic abnormalities contributes to disease progression remains poorly understood. Specifically, the biological and clinical consequences of concurrent high-risk alterations such as del(11q)/ATM-mutations and del(17p)/TP53-mutations have not been established.
Methods: We integrated next-generation sequencing (NGS) and clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 techniques to characterize the in vitro and in vivo effects of concurrent monoallelic or biallelic ATM and/or TP53 alterations in CLL prognosis, clonal evolution, and therapy response.
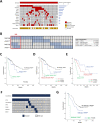
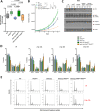
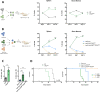
Results: Targeted sequencing analysis of the co-occurrence of high-risk alterations in 271 CLLs revealed that biallelic inactivation of both ATM and TP53 was mutually exclusive, whereas monoallelic del(11q) and TP53 alterations significantly co-occurred in a subset of CLL patients with a highly adverse clinical outcome. We determined the biological effects of combined del(11q), ATM and/or TP53 mutations in CRISPR/Cas9-edited CLL cell lines. Our results showed that the combination of monoallelic del(11q) and TP53 mutations in CLL cells led to a clonal advantage in vitro and in in vivo clonal competition experiments, whereas CLL cells harboring biallelic ATM and TP53 loss failed to compete in in vivo xenotransplants. Furthermore, we demonstrated that CLL cell lines harboring del(11q) and TP53 mutations show only partial responses to B cell receptor signaling inhibitors, but may potentially benefit from ATR inhibition.
Conclusions: Our work highlights that combined monoallelic del(11q) and TP53 alterations coordinately contribute to clonal advantage and shorter overall survival in CLL.
Keywords: CRISPR/Cas9 system; TP53 gene; biomarkers; chromosomal abnormality; chronic lymphocytic leukemia; next-generation sequencing.
© 2021 The Authors. Clinical and Translational Medicine published by John Wiley & Sons Australia, Ltd on behalf of Shanghai Institute of Clinical Bioinformatics.
Conflict of interest statement
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Figures


Similar articles
-
Biological significance of monoallelic and biallelic BIRC3 loss in del(11q) chronic lymphocytic leukemia progression.Blood Cancer J. 2021 Jul 9;11(7):127. doi: 10.1038/s41408-021-00520-5. Blood Cancer J. 2021. PMID: 34244476 Free PMC article.
-
CRISPR/Cas9-generated models uncover therapeutic vulnerabilities of del(11q) CLL cells to dual BCR and PARP inhibition.Leukemia. 2020 Jun;34(6):1599-1612. doi: 10.1038/s41375-020-0714-3. Epub 2020 Jan 23. Leukemia. 2020. PMID: 31974435 Free PMC article.
-
Survival of Del17p CLL Depends on Genomic Complexity and Somatic Mutation.Clin Cancer Res. 2017 Feb 1;23(3):735-745. doi: 10.1158/1078-0432.CCR-16-0594. Epub 2016 Aug 8. Clin Cancer Res. 2017. PMID: 27503198 Free PMC article.
-
The role of ATM mutations and 11q deletions in disease progression in chronic lymphocytic leukemia.Leuk Lymphoma. 2014 Jun;55(6):1227-39. doi: 10.3109/10428194.2013.829919. Epub 2013 Sep 12. Leuk Lymphoma. 2014. PMID: 23906020 Review.
-
The Heterogeneity of 13q Deletions in Chronic Lymphocytic Leukemia: Diagnostic Challenges and Clinical Implications.Genes (Basel). 2025 Feb 22;16(3):252. doi: 10.3390/genes16030252. Genes (Basel). 2025. PMID: 40149404 Free PMC article. Review.
Cited by
-
High-Throughput CRISPR Screening in Hematological Neoplasms.Cancers (Basel). 2022 Jul 25;14(15):3612. doi: 10.3390/cancers14153612. Cancers (Basel). 2022. PMID: 35892871 Free PMC article. Review.
-
The Evolving Landscape of Chronic Lymphocytic Leukemia on Diagnosis, Prognosis and Treatment.Diagnostics (Basel). 2021 May 10;11(5):853. doi: 10.3390/diagnostics11050853. Diagnostics (Basel). 2021. PMID: 34068813 Free PMC article. Review.
-
SAMHD1 dysfunction impairs DNA damage response and increases sensitivity to PARP inhibition in chronic lymphocytic leukemia.Sci Rep. 2025 Mar 26;15(1):10446. doi: 10.1038/s41598-025-93629-7. Sci Rep. 2025. PMID: 40140468 Free PMC article.
-
Mouse models of chronic lymphocytic leukemia and Richter transformation: what we have learnt and what we are missing.Front Immunol. 2024 Jun 6;15:1376660. doi: 10.3389/fimmu.2024.1376660. eCollection 2024. Front Immunol. 2024. PMID: 38903501 Free PMC article. Review.
-
Utilization of CRISPR-Mediated Tools for Studying Functional Genomics in Hematological Malignancies: An Overview on the Current Perspectives, Challenges, and Clinical Implications.Front Genet. 2022 Jan 28;12:767298. doi: 10.3389/fgene.2021.767298. eCollection 2021. Front Genet. 2022. PMID: 35154242 Free PMC article. Review.
References
-
- Chiorazzi N, Rai KR, Ferrarini M. Chronic lymphocytic leukemia. N Engl J Med. 2005;352(8):804–815. - PubMed
-
- Hallek M, Cheson BD, Catovsky D, et al. iwCLL guidelines for diagnosis, indications for treatment, response assessment, and supportive management of CLL. Blood. 2018;131(25):2745–2760. - PubMed
-
- Döhner H, Stilgenbauer S, Benner A, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343(26):1910–1916. - PubMed
-
- Puente XS, Beà S, Valdés‐Mas R, et al. Non‐coding recurrent mutations in chronic lymphocytic leukaemia. Nature. 2015;526(7574):519–524. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
