Acyl Transfer Catalytic Activity in De Novo Designed Protein with N-Terminus of α-Helix As Oxyanion-Binding Site
- PMID: 33635059
- PMCID: PMC8012002
- DOI: 10.1021/jacs.0c10053
Acyl Transfer Catalytic Activity in De Novo Designed Protein with N-Terminus of α-Helix As Oxyanion-Binding Site
Abstract
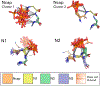
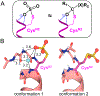
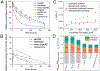
The design of catalytic proteins with functional sites capable of specific chemistry is gaining momentum and a number of artificial enzymes have recently been reported, including hydrolases, oxidoreductases, retro-aldolases, and others. Our goal is to develop a peptide ligase for robust catalysis of amide bond formation that possesses no stringent restrictions to the amino acid composition at the ligation junction. We report here the successful completion of the first step in this long-term project by building a completely de novo protein with predefined acyl transfer catalytic activity. We applied a minimalist approach to rationally design an oxyanion hole within a small cavity that contains an adjacent thiol nucleophile. The N-terminus of the α-helix with unpaired hydrogen-bond donors was exploited as a structural motif to stabilize negatively charged tetrahedral intermediates in nucleophilic addition-elimination reactions at the acyl group. Cysteine acting as a principal catalytic residue was introduced at the second residue position of the α-helix N-terminus in a designed three-α-helix protein based on structural informatics prediction. We showed that this minimal set of functional elements is sufficient for the emergence of catalytic activity in a de novo protein. Using peptide-αthioesters as acyl-donors, we demonstrated their catalyzed amidation concomitant with hydrolysis and proved that the environment at the catalytic site critically influences the reaction outcome. These results represent a promising starting point for the development of efficient catalysts for protein labeling, conjugation, and peptide ligation.
Figures




References
-
- Zanghellini A De Novo Computational Enzyme Design. Curr. Opin. Biotechnol 2014, 29, 132–138. - PubMed
-
- Kries H; Blomberg R; Hilvert D De Novo Enzymes by Computational Design. Curr. Opin. Chem. Biol 2013, 17, 221–228. - PubMed
-
- Hilvert D Design of Protein Catalysts. Annu. Rev. Biochem 2013, 82, 447–470. - PubMed
-
- Röthlisberger D; Khersonsky O; Wollacott AM; Jiang L; DeChancie J; Betker J; Gallaher JL; Althoff EA; Zanghellini A; Dym O; Albeck S; Houk KN; Tawfik DS; Baker D Kemp Elimination Catalysts by Computational Enzyme Design. Nature 2008, 453, 190–195. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

