Hematopoietic stem and progenitor cells are present in healthy gingiva tissue
- PMID: 33635312
- PMCID: PMC7923695
- DOI: 10.1084/jem.20200737
Hematopoietic stem and progenitor cells are present in healthy gingiva tissue
Abstract
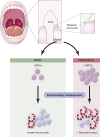
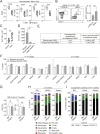
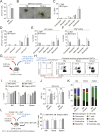
Hematopoietic stem cells reside in the bone marrow, where they generate the effector cells that drive immune responses. However, in response to inflammation, some hematopoietic stem and progenitor cells (HSPCs) are recruited to tissue sites and undergo extramedullary hematopoiesis. Contrasting with this paradigm, here we show residence and differentiation of HSPCs in healthy gingiva, a key oral barrier in the absence of overt inflammation. We initially defined a population of gingiva monocytes that could be locally maintained; we subsequently identified not only monocyte progenitors but also diverse HSPCs within the gingiva that could give rise to multiple myeloid lineages. Gingiva HSPCs possessed similar differentiation potentials, reconstitution capabilities, and heterogeneity to bone marrow HSPCs. However, gingival HSPCs responded differently to inflammatory insults, responding to oral but not systemic inflammation. Combined, we highlight a novel pathway of myeloid cell development at a healthy barrier, defining a gingiva-specific HSPC network that supports generation of a proportion of the innate immune cells that police this barrier.
© 2021 Krishnan et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures





References
-
- Bain, C.C., Scott C.L., Uronen-Hansson H., Gudjonsson S., Jansson O., Grip O., Guilliams M., Malissen B., Agace W.W., and Mowat A.M.. 2013. Resident and pro-inflammatory macrophages in the colon represent alternative context-dependent fates of the same Ly6Chi monocyte precursors. Mucosal Immunol. 6:498–510. 10.1038/mi.2012.89 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

