Identification of SARS-CoV-2-specific immune alterations in acutely ill patients
- PMID: 33635833
- PMCID: PMC8262478
- DOI: 10.1172/JCI145853
Identification of SARS-CoV-2-specific immune alterations in acutely ill patients
Abstract
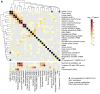
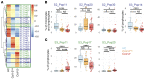
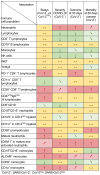
Dysregulated immune profiles have been described in symptomatic patients infected with SARS-CoV-2. Whether the reported immune alterations are specific to SARS-CoV-2 infection or also triggered by other acute illnesses remains unclear. We performed flow cytometry analysis on fresh peripheral blood from a consecutive cohort of (a) patients hospitalized with acute SARS-CoV-2 infection, (b) patients of comparable age and sex hospitalized for another acute disease (SARS-CoV-2 negative), and (c) healthy controls. Using both data-driven and hypothesis-driven analyses, we found several dysregulations in immune cell subsets (e.g., decreased proportion of T cells) that were similarly associated with acute SARS-CoV-2 infection and non-COVID-19-related acute illnesses. In contrast, we identified specific differences in myeloid and lymphocyte subsets that were associated with SARS-CoV-2 status (e.g., elevated proportion of ICAM-1+ mature/activated neutrophils, ALCAM+ monocytes, and CD38+CD8+ T cells). A subset of SARS-CoV-2-specific immune alterations correlated with disease severity, disease outcome at 30 days, and mortality. Our data provide an understanding of the immune dysregulation specifically associated with SARS-CoV-2 infection among acute care hospitalized patients. Our study lays the foundation for the development of specific biomarkers to stratify SARS-CoV-2-positive patients at risk of unfavorable outcomes and to uncover candidate molecules to investigate from a therapeutic perspective.
Keywords: Adaptive immunity; COVID-19; Innate immunity.
Conflict of interest statement
Figures





Similar articles
-
Lymphocyte Subset Alteration and Monocyte CD4 Expression Reduction in Patients with Severe COVID-19.Viral Immunol. 2021 Jun;34(5):342-351. doi: 10.1089/vim.2020.0166. Epub 2020 Nov 23. Viral Immunol. 2021. PMID: 33264073
-
Dynamic SARS-CoV-2-Specific Immunity in Critically Ill Patients With Hypertension.Front Immunol. 2020 Dec 10;11:596684. doi: 10.3389/fimmu.2020.596684. eCollection 2020. Front Immunol. 2020. PMID: 33362779 Free PMC article.
-
Disease-duration based comparison of subsets of immune cells in SARS CoV-2 infected patients presenting with mild or severe symptoms identifies prognostic markers for severity.Immun Inflamm Dis. 2021 Jun;9(2):419-434. doi: 10.1002/iid3.402. Epub 2021 Jan 16. Immun Inflamm Dis. 2021. PMID: 33452858 Free PMC article.
-
Comprehensive Analysis of the ILCs and Unconventional T Cells in Virus Infection: Profiling and Dynamics Associated with COVID-19 Disease for a Future Monitoring System and Therapeutic Opportunities.Cells. 2022 Feb 4;11(3):542. doi: 10.3390/cells11030542. Cells. 2022. PMID: 35159352 Free PMC article. Review.
-
Alterations in lymphocyte subsets and monocytes in patients diagnosed with SARS-CoV-2 pneumonia: a mini review of the literature.Eur Rev Med Pharmacol Sci. 2021 Aug;25(15):5057-5062. doi: 10.26355/eurrev_202108_26463. Eur Rev Med Pharmacol Sci. 2021. PMID: 34355378 Review.
Cited by
-
Plasmacytoid dendritic cells during COVID-19: Ally or adversary?Cell Rep. 2022 Jul 26;40(4):111148. doi: 10.1016/j.celrep.2022.111148. Epub 2022 Jul 14. Cell Rep. 2022. PMID: 35858624 Free PMC article. Review.
-
Evaluating Humoral Immunity against SARS-CoV-2: Validation of a Plaque-Reduction Neutralization Test and a Multilaboratory Comparison of Conventional and Surrogate Neutralization Assays.Microbiol Spectr. 2021 Dec 22;9(3):e0088621. doi: 10.1128/Spectrum.00886-21. Epub 2021 Nov 17. Microbiol Spectr. 2021. PMID: 34787495 Free PMC article.
-
Proteomic and Metabolomic Characterization of SARS-CoV-2-Infected Cynomolgus Macaque at Early Stage.Front Immunol. 2022 Jul 12;13:954121. doi: 10.3389/fimmu.2022.954121. eCollection 2022. Front Immunol. 2022. PMID: 35903092 Free PMC article.
-
The Initial COVID-19 Reliable Interactive DNA Methylation Markers and Biological Implications.Biology (Basel). 2024 Apr 7;13(4):245. doi: 10.3390/biology13040245. Biology (Basel). 2024. PMID: 38666857 Free PMC article.
-
COVID-19: Using high-throughput flow cytometry to dissect clinical heterogeneity.Cytometry A. 2023 Feb;103(2):117-126. doi: 10.1002/cyto.a.24516. Epub 2021 Nov 22. Cytometry A. 2023. PMID: 34811890 Free PMC article. Review.
References
-
- Onder G, et al. Case-fatality rate and characteristics of patients dying in relation to COVID-19 in Italy. JAMA. 2020;323(18):1775–1776. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

