Power analysis of transcriptome-wide association study: Implications for practical protocol choice
- PMID: 33635859
- PMCID: PMC7946362
- DOI: 10.1371/journal.pgen.1009405
Power analysis of transcriptome-wide association study: Implications for practical protocol choice
Abstract
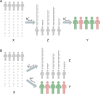
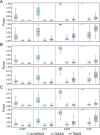
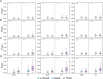
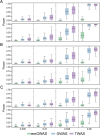
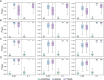
The transcriptome-wide association study (TWAS) has emerged as one of several promising techniques for integrating multi-scale 'omics' data into traditional genome-wide association studies (GWAS). Unlike GWAS, which associates phenotypic variance directly with genetic variants, TWAS uses a reference dataset to train a predictive model for gene expressions, which allows it to associate phenotype with variants through the mediating effect of expressions. Although effective, this core innovation of TWAS is poorly understood, since the predictive accuracy of the genotype-expression model is generally low and further bounded by expression heritability. This raises the question: to what degree does the accuracy of the expression model affect the power of TWAS? Furthermore, would replacing predictions with actual, experimentally determined expressions improve power? To answer these questions, we compared the power of GWAS, TWAS, and a hypothetical protocol utilizing real expression data. We derived non-centrality parameters (NCPs) for linear mixed models (LMMs) to enable closed-form calculations of statistical power that do not rely on specific protocol implementations. We examined two representative scenarios: causality (genotype contributes to phenotype through expression) and pleiotropy (genotype contributes directly to both phenotype and expression), and also tested the effects of various properties including expression heritability. Our analysis reveals two main outcomes: (1) Under pleiotropy, the use of predicted expressions in TWAS is superior to actual expressions. This explains why TWAS can function with weak expression models, and shows that TWAS remains relevant even when real expressions are available. (2) GWAS outperforms TWAS when expression heritability is below a threshold of 0.04 under causality, or 0.06 under pleiotropy. Analysis of existing publications suggests that TWAS has been misapplied in place of GWAS, in situations where expression heritability is low.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

