Computational screening of potential glioma-related genes and drugs based on analysis of GEO dataset and text mining
- PMID: 33635875
- PMCID: PMC7909668
- DOI: 10.1371/journal.pone.0247612
Computational screening of potential glioma-related genes and drugs based on analysis of GEO dataset and text mining
Abstract
Background: Considering the high invasiveness and mortality of glioma as well as the unclear key genes and signaling pathways involved in the development of gliomas, there is a strong need to find potential gene biomarkers and available drugs.
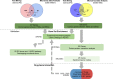
Methods: Eight glioma samples and twelve control samples were analyzed on the GSE31095 datasets, and differentially expressed genes (DEGs) were obtained via the R software. The related glioma genes were further acquired from the text mining. Additionally, Venny program was used to screen out the common genes of the two gene sets and DAVID analysis was used to conduct the corresponding gene ontology analysis and cell signal pathway enrichment. We also constructed the protein interaction network of common genes through STRING, and selected the important modules for further drug-gene analysis. The existing antitumor drugs that targeted these module genes were screened to explore their efficacy in glioma treatment.
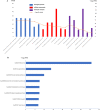
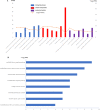
Results: The gene set obtained from text mining was intersected with the previously obtained DEGs, and 128 common genes were obtained. Through the functional enrichment analysis of the identified 128 DEGs, a hub gene module containing 25 genes was obtained. Combined with the functional terms in GSE109857 dataset, some overlap of the enriched function terms are both in GSE31095 and GSE109857. Finally, 4 antitumor drugs were identified through drug-gene interaction analysis.
Conclusions: In this study, we identified that two potential genes and their corresponding four antitumor agents could be used as targets and drugs for glioma exploration.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Identification of Hub Gene GRIN1 Correlated with Histological Grade and Prognosis of Glioma by Weighted Gene Coexpression Network Analysis.Biomed Res Int. 2021 Nov 19;2021:4542995. doi: 10.1155/2021/4542995. eCollection 2021. Biomed Res Int. 2021. PMID: 34840971 Free PMC article.
-
Integrated bioinformatics analysis for the screening of hub genes and therapeutic drugs in ovarian cancer.J Ovarian Res. 2020 Jan 27;13(1):10. doi: 10.1186/s13048-020-0613-2. J Ovarian Res. 2020. PMID: 31987036 Free PMC article.
-
The promising novel biomarkers and candidate small molecule drugs in lower-grade glioma: Evidence from bioinformatics analysis of high-throughput data.J Cell Biochem. 2019 Sep;120(9):15106-15118. doi: 10.1002/jcb.28773. Epub 2019 Apr 24. J Cell Biochem. 2019. PMID: 31020692
-
Identification of hub genes in placental dysfunction and recurrent pregnancy loss through transcriptome data mining: A meta-analysis.Taiwan J Obstet Gynecol. 2024 May;63(3):297-306. doi: 10.1016/j.tjog.2024.01.035. Taiwan J Obstet Gynecol. 2024. PMID: 38802191 Review.
-
Using neurofibromatosis-1 to better understand and treat pediatric low-grade glioma.J Child Neurol. 2008 Oct;23(10):1186-94. doi: 10.1177/0883073808321061. J Child Neurol. 2008. PMID: 18952585 Free PMC article. Review.
Cited by
-
Integrated Transcriptome Profiling Identifies Prognostic Hub Genes as Therapeutic Targets of Glioblastoma: Evidenced by Bioinformatics Analysis.ACS Omega. 2022 Jun 22;7(26):22531-22550. doi: 10.1021/acsomega.2c01820. eCollection 2022 Jul 5. ACS Omega. 2022. PMID: 35811900 Free PMC article.
-
Systematic Review of Molecular Targeted Therapies for Adult-Type Diffuse Glioma: An Analysis of Clinical and Laboratory Studies.Int J Mol Sci. 2023 Jun 21;24(13):10456. doi: 10.3390/ijms241310456. Int J Mol Sci. 2023. PMID: 37445633 Free PMC article.
-
A Hybrid Protocol for Finding Novel Gene Targets for Various Diseases Using Microarray Expression Data Analysis and Text Mining.Methods Mol Biol. 2022;2496:41-70. doi: 10.1007/978-1-0716-2305-3_3. Methods Mol Biol. 2022. PMID: 35713858
-
In-silico identification and functional characterization of common genes associated with type 2 diabetes and hypertension.Heliyon. 2024 Aug 21;10(16):e36546. doi: 10.1016/j.heliyon.2024.e36546. eCollection 2024 Aug 30. Heliyon. 2024. PMID: 39262940 Free PMC article.
-
Inhibitory Effect of miR-339-5p on Glioma through PTP4A1/HMGB1 Pathway.Dis Markers. 2022 Jul 15;2022:2231195. doi: 10.1155/2022/2231195. eCollection 2022. Dis Markers. 2022. PMID: 35872698 Free PMC article.
References
-
- Meyer MA. Malignant gliomas in adults. New England Journal of Medicine. 2008;359(17):1850. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

