Osteoclasts recycle via osteomorphs during RANKL-stimulated bone resorption
- PMID: 33636130
- PMCID: PMC7938889
- DOI: 10.1016/j.cell.2021.02.002
Osteoclasts recycle via osteomorphs during RANKL-stimulated bone resorption
Erratum in
-
Osteoclasts recycle via osteomorphs during RANKL-stimulated bone resorption.Cell. 2021 Apr 1;184(7):1940. doi: 10.1016/j.cell.2021.03.010. Cell. 2021. PMID: 33798441 Free PMC article. No abstract available.
Abstract
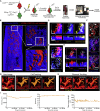
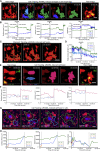
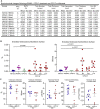
Osteoclasts are large multinucleated bone-resorbing cells formed by the fusion of monocyte/macrophage-derived precursors that are thought to undergo apoptosis once resorption is complete. Here, by intravital imaging, we reveal that RANKL-stimulated osteoclasts have an alternative cell fate in which they fission into daughter cells called osteomorphs. Inhibiting RANKL blocked this cellular recycling and resulted in osteomorph accumulation. Single-cell RNA sequencing showed that osteomorphs are transcriptionally distinct from osteoclasts and macrophages and express a number of non-canonical osteoclast genes that are associated with structural and functional bone phenotypes when deleted in mice. Furthermore, genetic variation in human orthologs of osteomorph genes causes monogenic skeletal disorders and associates with bone mineral density, a polygenetic skeletal trait. Thus, osteoclasts recycle via osteomorphs, a cell type involved in the regulation of bone resorption that may be targeted for the treatment of skeletal diseases.
Keywords: RANKL; cell fission; cellular recycling; denosumab; macrophage; osteoclast; osteomorph; osteoporosis; osteoprotegerin; skeletal dysplasia.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests F.H.E. and S.S. are employees and shareholders of BioVinc LLC, who has licensed patents for fluorescent probe compositions used in this work. P.I.C. has grant funding from Amgen. Other authors have no competing financial interests.
Figures




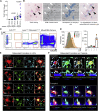

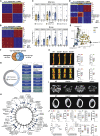
Comment in
-
Bone resorption goes green.Cell. 2021 Mar 4;184(5):1137-1139. doi: 10.1016/j.cell.2021.02.023. Epub 2021 Feb 25. Cell. 2021. PMID: 33636131 Free PMC article.
References
-
- Anastasilakis A.D., Polyzos S.A., Makras P., Aubry-Rozier B., Kaouri S., Lamy O. Clinical Features of 24 Patients With Rebound-Associated Vertebral Fractures After Denosumab Discontinuation: Systematic Review and Additional Cases. J. Bone Miner. Res. 2017;32:1291–1296. - PubMed
-
- Anastasilakis A.D., Yavropoulou M.P., Makras P., Sakellariou G.T., Papadopoulou F., Gerou S., Papapoulos S.E. Increased osteoclastogenesis in patients with vertebral fractures following discontinuation of denosumab treatment. Eur. J. Endocrinol. 2017;176:677–683. - PubMed
-
- Anderson J.M. Multinucleated giant cells. Curr. Opin. Hematol. 2000;7:40–47. - PubMed
-
- Arnett T.R., Dempster D.W. Effect of pH on bone resorption by rat osteoclasts in vitro. Endocrinology. 1986;119:119–124. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

