Immune subtyping for pancreatic cancer with implication in clinical outcomes and improving immunotherapy
- PMID: 33637086
- PMCID: PMC7908647
- DOI: 10.1186/s12935-021-01824-z
Immune subtyping for pancreatic cancer with implication in clinical outcomes and improving immunotherapy
Abstract
Background: Emerging evidence has shown that intra-tumor immune features are associated with response to immune checkpoint blockade (ICB) therapy. Accordingly, patient stratification is needed for identifying target patients and designing strategies to improve the efficacy of ICB therapy. We aimed to depict the specific immune features of patients with pancreatic cancer and explore the implication of immune diversity in prognostic prediction and individualized immunotherapy.
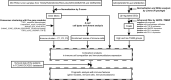
Methods: From transcriptional profiles of 383 tumor samples in TCGA, ICGC, and GEO database, robust immune subtypes which had different response immunotherapy, including ICB therapy, were identified by consensus clustering with five gene modules. DEGs analysis and tumor microarray were used to screen and demonstrate potential targets for improving ICB therapy.
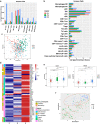
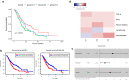
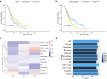
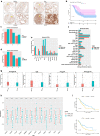
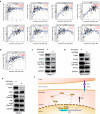
Results: Three subtypes of pancreatic cancer, namely cluster 1-3 (C1-C3), characterized with distinct immune features and prognosis, were generated. Of that, subtype C1 was an immune-cold type in lack of immune regulators, subtype C2, with an immunosuppression-dominated phenotype characterized by robust TGFβ signaling and stromal reaction, showed the worst prognosis, subtype C3 was an immune-hot type, with massive immune cell infiltration and in abundance of immune regulators. The disparity of immune features uncovered the discrepant applicability of anti-PD-1/PD-L1 therapy and potential sensitivity to other alternative immunotherapy for each subtype. Patients in C3 were more suitable for anti-PD-1/PD-L1 therapy, while patients in the other two clusters may need combined strategies targeted on other immune checkpoints or oncogenic pathways. A promising target for improving anti-PD-1/PD-L1 treatment, TGM2, was screened out and its role in the regulation of PD-L1 was investigated for the first time.
Conclusion: Collectively, immune features of pancreatic cancer contribute to distinct immunosuppressive mechanisms that are responsible for individualized immunotherapy. Despite pancreatic cancer being considered as a poor immunogenic cancer type, the derived immune subtypes may have implications in tailored designing of immunotherapy for the patients. TGM2 has potential synergistic roles with ICB therapy.
Keywords: Heterogeneity; Immune cell; Immune checkpoints; Immunotherapy; Pancreatic cancer; Transglutaminase 2.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

