Identification and characterization of heteroclitic peptides in TCR-binding positions with improved HLA-binding efficacy
- PMID: 33637105
- PMCID: PMC7913412
- DOI: 10.1186/s12967-021-02757-x
Identification and characterization of heteroclitic peptides in TCR-binding positions with improved HLA-binding efficacy
Abstract
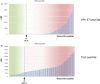
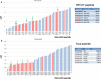
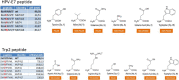
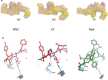
The antigenicity as well as the immunogenicity of tumor associated antigens (TAAs) may need to be potentiated in order to break the immunological tolerance. To this aim, heteroclitic peptides were designed introducing specific substitutions in the residue at position 4 (p4) binding to TCR. The effect of such modifications also on the affinity to the major histocompatibility class I (MHC-I) molecule was assessed. The Trp2 antigen, specific for the mouse melanoma B16F10 cells, as well as the HPV-E7 antigen, specific for the TC1 tumor cell lines, were used as models. Affinity of such heteroclitic peptides to HLA was predicted by bioinformatics tools and the most promising ones were validated by structural conformational and HLA binding analyses. Overall, we demonstrated that TAAs modified at the TCR-binding p4 residue are predicted to have higher affinity to MHC-I molecules. Experimental evaluation confirms the stronger binding, suggesting that this strategy may be very effective for designing new vaccines with improved antigenic efficacy.
Conflict of interest statement
The authors declare no potential conflicts of interest.
Figures
Similar articles
-
MHC-Optimized Peptide Scaffold for Improved Antigen Presentation and Anti-Tumor Response.Front Immunol. 2021 Oct 20;12:769799. doi: 10.3389/fimmu.2021.769799. eCollection 2021. Front Immunol. 2021. PMID: 34745146 Free PMC article.
-
Induction of cytotoxic T-cell responses against immunoglobulin V region-derived peptides modified at human leukocyte antigen-A2 binding residues.Blood. 2001 Nov 15;98(10):2999-3005. doi: 10.1182/blood.v98.10.2999. Blood. 2001. PMID: 11698283
-
Amino acid substitutions at position 97 in HLA-A2 segregate cytolysis from cytokine release in MART-1/Melan-A peptide AAGIGILTV-specific cytotoxic T lymphocytes.Eur J Immunol. 1996 Nov;26(11):2613-23. doi: 10.1002/eji.1830261112. Eur J Immunol. 1996. PMID: 8921947
-
MM-GBSA binding free energy decomposition and T cell receptor engineering.J Mol Recognit. 2010 Mar-Apr;23(2):142-52. doi: 10.1002/jmr.1005. J Mol Recognit. 2010. PMID: 20151417 Review.
-
Based on HLA-DR beta1* allele binding specificities, striking differences in distance and TCR Contacting Residue Orientation can be observed in modified protection-inducing malarial synthetic peptides.Curr Med Chem. 2005;12(24):2849-65. doi: 10.2174/092986705774454733. Curr Med Chem. 2005. PMID: 16305475 Review.
Cited by
-
Proceedings of the Online Conference "Vaccines and Vaccination during and Post COVID Pandemics" (7-9 December 2022).Vaccines (Basel). 2023 Jun 29;11(7):1175. doi: 10.3390/vaccines11071175. Vaccines (Basel). 2023. PMID: 37514990 Free PMC article.
-
Long-term memory T cells as preventive anticancer immunity elicited by TuA-derived heteroclitic peptides.J Transl Med. 2021 Dec 24;19(1):526. doi: 10.1186/s12967-021-03194-6. J Transl Med. 2021. PMID: 34952611 Free PMC article.
-
Recent Findings on Therapeutic Cancer Vaccines: An Updated Review.Biomolecules. 2024 Apr 21;14(4):503. doi: 10.3390/biom14040503. Biomolecules. 2024. PMID: 38672519 Free PMC article. Review.
-
Short Peptides as Powerful Arsenal for Smart Fighting Cancer.Cancers (Basel). 2024 Sep 24;16(19):3254. doi: 10.3390/cancers16193254. Cancers (Basel). 2024. PMID: 39409876 Free PMC article. Review.
-
Peptide-based vaccine for cancer therapies.Front Immunol. 2023 Aug 16;14:1210044. doi: 10.3389/fimmu.2023.1210044. eCollection 2023. Front Immunol. 2023. PMID: 37654484 Free PMC article. Review.
References
-
- Parker KC, Bednarek MA, Hull LK, Utz U, Cunningham B, Zweerink HJ, Biddison WE, Coligan JE. Sequence motifs important for peptide binding to the human MHC class I molecule, HLA-A2. J Immunol. 1992;149:3580–3587. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

