Contagious Antibiotic Resistance: Plasmid Transfer among Bacterial Residents of the Zebrafish Gut
- PMID: 33637574
- PMCID: PMC8091013
- DOI: 10.1128/AEM.02735-20
Contagious Antibiotic Resistance: Plasmid Transfer among Bacterial Residents of the Zebrafish Gut
Abstract
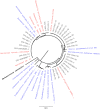
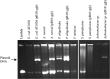
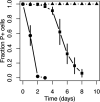
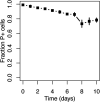
By characterizing the trajectories of antibiotic resistance gene transfer in bacterial communities such as the gut microbiome, we will better understand the factors that influence this spread of resistance. Our aim was to investigate the host network of a multidrug resistance broad-host-range plasmid in the culturable gut microbiome of zebrafish. This was done through in vitro and in vivo conjugation experiments with Escherichia coli as the donor of the plasmid pB10::gfp When this donor was mixed with the extracted gut microbiome, only transconjugants of Aeromonas veronii were detected. In separate matings between the same donor and four prominent isolates from the gut microbiome, the plasmid transferred to two of these four isolates, A. veronii and Plesiomonas shigelloides, but not to Shewanella putrefaciens and Vibrio mimicus When these A. veronii and P. shigelloides transconjugants were the donors in matings with the same four isolates, the plasmid now also transferred from A. veronii to S. putrefaciensP. shigelloides was unable to donate the plasmid, and V. mimicus was unable to acquire it. Finally, when the E. coli donor was added in vivo to zebrafish through their food, plasmid transfer was observed in the gut, but only to Achromobacter, a rare member of the gut microbiome. This work shows that the success of plasmid-mediated antibiotic resistance spread in a gut microbiome depends on the donor-recipient species combinations and therefore their spatial arrangement. It also suggests that rare gut microbiome members should not be ignored as potential reservoirs of multidrug resistance plasmids from food.IMPORTANCE To understand how antibiotic resistance plasmids end up in human pathogens, it is crucial to learn how, where, and when they are transferred and maintained in members of bacterial communities such as the gut microbiome. To gain insight into the network of plasmid-mediated antibiotic resistance sharing in the gut microbiome, we investigated the transferability and maintenance of a multidrug resistance plasmid among the culturable bacteria of the zebrafish gut. We show that the success of plasmid-mediated antibiotic resistance spread in a gut microbiome can depend on which species are involved, as some are important nodes in the plasmid-host network and others are dead ends. Our findings also suggest that rare gut microbiome members should not be ignored as potential reservoirs of multidrug resistance plasmids from food.
Keywords: conjugation; gut microbiome; horizontal gene transfer; plasmid; plasmid network; plasmid persistence; zebrafish.
Copyright © 2021 American Society for Microbiology.
Figures

References
-
- Kåhrström CT. 2013. Entering a post-antibiotic era? Nat Rev Microbiol 11:146. 10.1038/nrmicro2983. - DOI
-
- World Health Organization. 2018. Global Antimicrobial Resistance Surveillance System (GLASS) report: early implementation 2017–2018. World Health Organization, Geneva, Switzerland.
-
- World Health Organization. 2014. Antimicrobial resistance: global report on surveillance. World Health Organization, Geneva, Switzerland.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

