Human airway mast cells proliferate and acquire distinct inflammation-driven phenotypes during type 2 inflammation
- PMID: 33637594
- PMCID: PMC8362933
- DOI: 10.1126/sciimmunol.abb7221
Human airway mast cells proliferate and acquire distinct inflammation-driven phenotypes during type 2 inflammation
Abstract
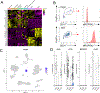
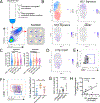
Mast cells (MCs) play a pathobiologic role in type 2 (T2) allergic inflammatory diseases of the airway, including asthma and chronic rhinosinusitis with nasal polyposis (CRSwNP). Distinct MC subsets infiltrate the airway mucosa in T2 disease, including subepithelial MCs expressing the proteases tryptase and chymase (MCTC) and epithelial MCs expressing tryptase without chymase (MCT). However, mechanisms underlying MC expansion and the transcriptional programs underlying their heterogeneity are poorly understood. Here, we use flow cytometry and single-cell RNA-sequencing (scRNA-seq) to conduct a comprehensive analysis of human MC hyperplasia in CRSwNP, a T2 cytokine-mediated inflammatory disease. We link discrete cell surface phenotypes to the distinct transcriptomes of CRSwNP MCT and MCTC, which represent polarized ends of a transcriptional gradient of nasal polyp MCs. We find a subepithelial population of CD38highCD117high MCs that is markedly expanded during T2 inflammation. These CD38highCD117high MCs exhibit an intermediate phenotype relative to the expanded MCT and MCTC subsets. CD38highCD117high MCs are distinct from circulating MC progenitors and are enriched for proliferation, which is markedly increased in CRSwNP patients with aspirin-exacerbated respiratory disease, a severe disease subset characterized by increased MC burden and elevated MC activation. We observe that MCs expressing a polyp MCT-like effector program are also found within the lung during fibrotic diseases and asthma, and further identify marked differences between MCTC in nasal polyps and skin. These results indicate that MCs display distinct inflammation-associated effector programs and suggest that in situ MC proliferation is a major component of MC hyperplasia in human T2 inflammation.
Copyright © 2021 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Competing interests
J.O.M. reports compensation for consulting services with Cellarity and Hovione. R.E.R. reports compensation for consulting services with Roche Genentech and Gossamer pharmaceuticals. N.A.B. reports compensation for consulting services for Regeneron. A.K.S. reports compensation for consulting and/or SAB membership from Merck, Honeycomb Biotechnologies, Cellarity, Repertoire Immune Medicines, Ocrhe Bio, Hovione, and Dahlia Biosciences.
Figures




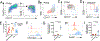
References
Publication types
MeSH terms
Grants and funding
- R01 HL128241/HL/NHLBI NIH HHS/United States
- R01 AI134989/AI/NIAID NIH HHS/United States
- K23 AI139352/AI/NIAID NIH HHS/United States
- T32 AI007306/AI/NIAID NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01 AI078908/AI/NIAID NIH HHS/United States
- R01 HL120952/HL/NHLBI NIH HHS/United States
- R01 HL136209/HL/NHLBI NIH HHS/United States
- U24 AI118672/AI/NIAID NIH HHS/United States
- R37 AI052353/AI/NIAID NIH HHS/United States
- R01 AI136041/AI/NIAID NIH HHS/United States
- K22 AI146281/AI/NIAID NIH HHS/United States
- U19 AI095219/AI/NIAID NIH HHS/United States
- U19 AI070535/AI/NIAID NIH HHS/United States
- P30 DK034854/DK/NIDDK NIH HHS/United States
- T32 GM087237/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

