Uncovering candidate genes involved in photosynthetic capacity using unexplored genetic variation in Spring Wheat
- PMID: 33638599
- PMCID: PMC8384606
- DOI: 10.1111/pbi.13568
Uncovering candidate genes involved in photosynthetic capacity using unexplored genetic variation in Spring Wheat
Abstract
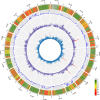
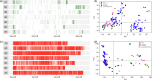
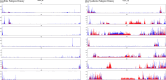
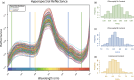
To feed an ever-increasing population we must leverage advances in genomics and phenotyping to harness the variation in wheat breeding populations for traits like photosynthetic capacity which remains unoptimized. Here we survey a diverse set of wheat germplasm containing elite, introgression and synthetic derivative lines uncovering previously uncharacterized variation. We demonstrate how strategic integration of exotic material alleviates the D genome genetic bottleneck in wheat, increasing SNP rate by 62% largely due to Ae. tauschii synthetic wheat donors. Across the panel, 67% of the Ae. tauschii donor genome is represented as introgressions in elite backgrounds. We show how observed genetic variation together with hyperspectral reflectance data can be used to identify candidate genes for traits relating to photosynthetic capacity using association analysis. This demonstrates the value of genomic methods in uncovering hidden variation in wheat and how that variation can assist breeding efforts and increase our understanding of complex traits.
Keywords: Aegilops Tauschii; Triticum aestivum; GWAS; capture sequencing; exotic material; hyperspectral reflectance.
© 2021 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Allen, A.M. , Winfield, M.O. , Burridge, A.J. , Downie, R.C. , Benbow, H.R. , Barker, G.L.A. , Wilkinson, P.A. et al. (2016) Characterization of a Wheat Breeders’ Array suitable for high‐throughput SNP genotyping of global accessions of hexaploid bread wheat (Triticum aestivum). Plant Biotechnol. J. 15, 390–401. - PMC - PubMed
-
- Alvarado, G. , López, M. , Vargas, M. , Pacheco, Á. , Rodríguez, F. , Burgueño, J. and Crossa, J. (2019). META‐R (Multi Environment Trail Analysis with R for Windows) Version 6.04.
-
- Araus, J.‐L. and Cairns, J.E. (2014) Field high‐throughput phenotyping: the new crop breeding frontier. Trends Plant Sci. 19, 52–61. - PubMed
-
- Bhusal, N. , Sharma, P. , Sareen, S. and Sarial, A.K. (2018) Mapping QTLs for chlorophyll content and chlorophyll fluorescence in wheat under heat stress. Biol. Plant. 62, 721–731.
-
- Blackburn, G.A. (2006) Hyperspectral remote sensing of plant pigments. J. Exp. Bot. 58, 855–867. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

