Gene Targeting Facilitated by Engineered Sequence-Specific Nucleases: Potential Applications for Crop Improvement
- PMID: 33638992
- PMCID: PMC8484935
- DOI: 10.1093/pcp/pcab034
Gene Targeting Facilitated by Engineered Sequence-Specific Nucleases: Potential Applications for Crop Improvement
Abstract
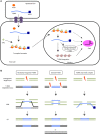
Humans are currently facing the problem of how to ensure that there is enough food to feed all of the world's population. Ensuring that the food supply is sufficient will likely require the modification of crop genomes to improve their agronomic traits. The development of engineered sequence-specific nucleases (SSNs) paved the way for targeted gene editing in organisms, including plants. SSNs generate a double-strand break (DSB) at the target DNA site in a sequence-specific manner. These DSBs are predominantly repaired via error-prone non-homologous end joining and are only rarely repaired via error-free homology-directed repair if an appropriate donor template is provided. Gene targeting (GT), i.e. the integration or replacement of a particular sequence, can be achieved with combinations of SSNs and repair donor templates. Although its efficiency is extremely low, GT has been achieved in some higher plants. Here, we provide an overview of SSN-facilitated GT in higher plants and discuss the potential of GT as a powerful tool for generating crop plants with desirable features.
Keywords: Crop; Gene targeting; Sequence-specific nuclease.
© The Author(s) 2021. Published by Oxford University Press on behalf of Japanese Society of Plant Physiologists.
Figures

Similar articles
-
Precise Genome Modification via Sequence-Specific Nucleases-Mediated Gene Targeting for Crop Improvement.Front Plant Sci. 2016 Dec 20;7:1928. doi: 10.3389/fpls.2016.01928. eCollection 2016. Front Plant Sci. 2016. PMID: 28066481 Free PMC article. Review.
-
Revisiting CRISPR/Cas-mediated crop improvement: Special focus on nutrition.J Biosci. 2020;45:137. J Biosci. 2020. PMID: 33361628
-
Genome editing for crop improvement: Challenges and opportunities.GM Crops Food. 2015;6(4):183-205. doi: 10.1080/21645698.2015.1129937. GM Crops Food. 2015. PMID: 26930114 Free PMC article.
-
Use of designer nucleases for targeted gene and genome editing in plants.Plant Biotechnol J. 2016 Feb;14(2):483-95. doi: 10.1111/pbi.12448. Epub 2015 Aug 11. Plant Biotechnol J. 2016. PMID: 26261084 Free PMC article. Review.
-
Towards mastering CRISPR-induced gene knock-in in plants: Survey of key features and focus on the model Physcomitrella patens.Methods. 2017 May 15;121-122:103-117. doi: 10.1016/j.ymeth.2017.04.024. Epub 2017 May 4. Methods. 2017. PMID: 28478103 Review.
Cited by
-
Precise and heritable gene targeting in rice using a sequential transformation strategy.Cell Rep Methods. 2023 Jan 17;3(1):100389. doi: 10.1016/j.crmeth.2022.100389. eCollection 2023 Jan 23. Cell Rep Methods. 2023. PMID: 36814841 Free PMC article.
-
Exploiting viral vectors to deliver genome editing reagents in plants.aBIOTECH. 2024 May 8;5(2):247-261. doi: 10.1007/s42994-024-00147-7. eCollection 2024 Jun. aBIOTECH. 2024. PMID: 38974861 Free PMC article. Review.
-
Simple promotion of Cas9 and Cas12a expression improves gene targeting via an all-in-one strategy.Front Plant Sci. 2024 Mar 13;15:1360925. doi: 10.3389/fpls.2024.1360925. eCollection 2024. Front Plant Sci. 2024. PMID: 38545386 Free PMC article.
-
Cas12a-mediated gene targeting by sequential transformation strategy in Arabidopsis thaliana.BMC Plant Biol. 2024 Jul 12;24(1):665. doi: 10.1186/s12870-024-05375-z. BMC Plant Biol. 2024. PMID: 38997669 Free PMC article.
-
Insights into the molecular mechanisms of CRISPR/Cas9-mediated gene targeting at multiple loci in Arabidopsis.Plant Physiol. 2022 Nov 28;190(4):2203-2216. doi: 10.1093/plphys/kiac431. Plant Physiol. 2022. PMID: 36106983 Free PMC article.
References
-
- Ainley W.M., Sastry-Dent L., Welter M.E., Murray M.G., Zeitler B., Amora R., et al. (2013) Trait stacking via targeted genome editing. Plant Biotechnol. J. 11: 1126–1134. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

