Interspecies Differences in Proteome Turnover Kinetics Are Correlated With Life Spans and Energetic Demands
- PMID: 33639418
- PMCID: PMC7950207
- DOI: 10.1074/mcp.RA120.002301
Interspecies Differences in Proteome Turnover Kinetics Are Correlated With Life Spans and Energetic Demands
Abstract
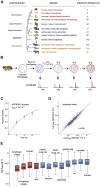
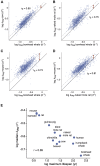
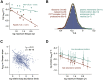
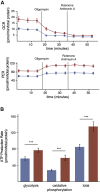
Cells continually degrade and replace damaged proteins. However, the high energetic demand of protein turnover generates reactive oxygen species that compromise the long-term health of the proteome. Thus, the relationship between aging, protein turnover, and energetic demand remains unclear. Here, we used a proteomic approach to measure rates of protein turnover within primary fibroblasts isolated from a number of species with diverse life spans including the longest-lived mammal, the bowhead whale. We show that organismal life span is negatively correlated with turnover rates of highly abundant proteins. In comparison with mice, cells from long-lived naked mole rats have slower rates of protein turnover, lower levels of ATP production, and reduced reactive oxygen species levels. Despite having slower rates of protein turnover, naked mole rat cells tolerate protein misfolding stress more effectively than mouse cells. We suggest that in lieu of a rapid constitutive turnover, long-lived species may have evolved more energetically efficient mechanisms for selective detection and clearance of damaged proteins.
Keywords: Protein turnove; aging; protein degradation; proteostasis; quantitative proteomics.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare no competing interests.
Figures




Similar articles
-
Cross-species Comparison of Proteome Turnover Kinetics.Mol Cell Proteomics. 2018 Apr;17(4):580-591. doi: 10.1074/mcp.RA117.000574. Epub 2018 Jan 10. Mol Cell Proteomics. 2018. PMID: 29321186 Free PMC article.
-
Proteome Turnover in the Spotlight: Approaches, Applications, and Perspectives.Mol Cell Proteomics. 2021;20:100016. doi: 10.1074/mcp.R120.002190. Epub 2020 Dec 7. Mol Cell Proteomics. 2021. PMID: 33556866 Free PMC article. Review.
-
Metabolic clues to salubrious longevity in the brain of the longest-lived rodent: the naked mole-rat.J Neurochem. 2015 Aug;134(3):538-50. doi: 10.1111/jnc.13149. Epub 2015 Jun 3. J Neurochem. 2015. PMID: 25940666
-
Comparative Study of Protein Aggregation Propensity and Mutation Tolerance Between Naked Mole-Rat and Mouse.Genome Biol Evol. 2022 May 3;14(5):evac057. doi: 10.1093/gbe/evac057. Genome Biol Evol. 2022. PMID: 35482036 Free PMC article.
-
The naked mole-rat response to oxidative stress: just deal with it.Antioxid Redox Signal. 2013 Oct 20;19(12):1388-99. doi: 10.1089/ars.2012.4911. Epub 2012 Dec 7. Antioxid Redox Signal. 2013. PMID: 23025341 Free PMC article. Review.
Cited by
-
Primary Cell Culture as a Model System for Evolutionary Molecular Physiology.Int J Mol Sci. 2024 Jul 19;25(14):7905. doi: 10.3390/ijms25147905. Int J Mol Sci. 2024. PMID: 39063147 Free PMC article. Review.
-
Fighting with Aging: The Secret for Keeping Health and Longevity of Naked Mole Rats.Aging Dis. 2024 Jan 11;16(1):137-45. doi: 10.14336/AD.2024.0109. Online ahead of print. Aging Dis. 2024. PMID: 38300643 Free PMC article. Review.
-
Metabolic mechanisms of species-specific developmental tempo.Dev Cell. 2024 Jul 8;59(13):1628-1639. doi: 10.1016/j.devcel.2024.05.027. Epub 2024 Jun 20. Dev Cell. 2024. PMID: 38906137 Free PMC article. Review.
-
Methionine Metabolism Is Down-Regulated in Heart of Long-Lived Mammals.Biology (Basel). 2022 Dec 14;11(12):1821. doi: 10.3390/biology11121821. Biology (Basel). 2022. PMID: 36552330 Free PMC article.
-
Naked mole-rat and Damaraland mole-rat exhibit lower respiration in mitochondria, cellular and organismal levels.Biochim Biophys Acta Bioenerg. 2022 Oct 1;1863(7):148582. doi: 10.1016/j.bbabio.2022.148582. Epub 2022 Jun 3. Biochim Biophys Acta Bioenerg. 2022. PMID: 35667393 Free PMC article.
References
-
- Sontag E.M., Samant R.S., Frydman J. Mechanisms and functions of spatial protein quality control. Annu. Rev. Biochem. 2017;86:97–122. - PubMed
-
- Pratt J.M., Petty J., Riba-Garcia I., Robertson D.H., Gaskell S.J., Oliver S.G., Beynon R.J. Dynamics of protein turnover, a missing dimension in proteomics. Mol. Cell. Proteomics. 2002;1:579–591. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

