Transcriptional Biomarkers in Oral Cancer: An Integrative Analysis and the Cancer Genome Atlas Validation
- PMID: 33639650
- PMCID: PMC8190349
- DOI: 10.31557/APJCP.2021.22.2.371
Transcriptional Biomarkers in Oral Cancer: An Integrative Analysis and the Cancer Genome Atlas Validation
Abstract
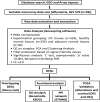
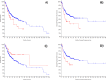
Objective: An impervious mortality rate in oral cancer (OC) to a certain extent explains the exigencies of precise biomarkers. Therefore, the study was intended to identify OC candidate biomarkers using samples of healthy normal tissues (N=335), adjacent normal tissues (N=93) and OC tissues (N=533) from online microarray data.
Methods: Differentially expressed genes (DEGs) were recognised through GeneSpring software (Fold change >4.0 and 'p' value.
Keywords: Biomarkers; expression profiling by array; integrative analysis; oral cancer; the cancer genome atlas.
Figures


Similar articles
-
An integrative analysis to enumerate candidate genes for clinical use in oral cancer.J Cancer Res Ther. 2022 Dec;18(Supplement):S182-S190. doi: 10.4103/jcrt.JCRT_1607_20. J Cancer Res Ther. 2022. PMID: 36510962
-
Integrated bioinformatics analysis for the screening of hub genes and therapeutic drugs in ovarian cancer.J Ovarian Res. 2020 Jan 27;13(1):10. doi: 10.1186/s13048-020-0613-2. J Ovarian Res. 2020. PMID: 31987036 Free PMC article.
-
Cancer Stem Cell based molecular predictors of tumor recurrence in Oral squamous cell carcinoma.Arch Oral Biol. 2019 Mar;99:92-106. doi: 10.1016/j.archoralbio.2019.01.002. Epub 2019 Jan 4. Arch Oral Biol. 2019. PMID: 30641296
-
Identification of a Gene Prognostic Signature for Oral Squamous Cell Carcinoma by RNA Sequencing and Bioinformatics.Biomed Res Int. 2021 Apr 1;2021:6657767. doi: 10.1155/2021/6657767. eCollection 2021. Biomed Res Int. 2021. PMID: 33869632 Free PMC article.
-
Molecular Biomarkers Related to Oral Carcinoma: Clinical Trial Outcome Evaluation in a Literature Review.Dis Markers. 2019 Mar 25;2019:8040361. doi: 10.1155/2019/8040361. eCollection 2019. Dis Markers. 2019. PMID: 31019584 Free PMC article.
Cited by
-
AEGAN-Pathifier: a data augmentation method to improve cancer classification for imbalanced gene expression data.BMC Bioinformatics. 2024 Dec 27;25(1):392. doi: 10.1186/s12859-024-06013-z. BMC Bioinformatics. 2024. PMID: 39731019 Free PMC article.
References
-
- Ademuyiwa FO, Miller A, O’Connor T, et al. The effects of oncotype DX recurrence scores on chemotherapy utilization in a multi-institutional breast cancer cohort. Breast Cancer Res Treat. 2011;126:797–802. - PubMed
-
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

