Measurement of gene amplifications related to drug resistance in Plasmodium falciparum using droplet digital PCR
- PMID: 33639924
- PMCID: PMC7916280
- DOI: 10.1186/s12936-021-03659-5
Measurement of gene amplifications related to drug resistance in Plasmodium falciparum using droplet digital PCR
Abstract
Background: Copy number variations (CNVs) of the Plasmodium falciparum multidrug resistance 1 (pfmdr1), P. falciparum plasmepsin2 (pfplasmepsin2) and P. falciparum GTP cyclohydrolase 1 (pfgch1) genes are associated with anti-malarial drug resistance in P. falciparum malaria. Droplet digital PCR (ddPCR) assays have been developed for accurate assessment of CNVs in several human genes. The aim of the present study was to develop and validate ddPCR assays for detection of the CNVs of P. falciparum genes associated with resistance to anti-malarial drugs.
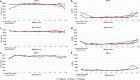
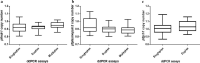
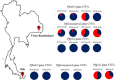
Methods: A multiplex ddPCR assay was developed to detect the CNVs in the pfmdr1 and pfplasmepsin2 genes, while a duplex ddPCR assay was developed to detect CNV in the pfgch1 gene. The gene copy number (GCN) quantification limit, as well as the accuracy and precision of the ddPCR assays were determined and compared to conventional quantitative PCR (qPCR). In order to reduce the cost of testing, a multiplex ddPCR assay of two target genes, pfmdr1 and pfplasmepsin2, was validated. In addition, the CNVs of genes of field samples collected from Thailand from 2015 to 2019 (n = 84) were assessed by ddPCR and results were compared to qPCR as the reference assay.
Results: There were no significant differences between the GCN results obtained from uniplex and multiplex ddPCR assays for detection of CNVs in the pfmdr1 and pfplasmepsin2 genes (p = 0.363 and 0.330, respectively). Based on the obtained gene copy number quantification limit, the accuracy and percent relative standard deviation (%RSD) value of the multiplex ddPCR assay were 95% and 5%, respectively, for detection of the CNV of the pfmdr1 gene, and 91% and 5% for detection of the CNV of the pfplasmepsin2 gene. There was no significant difference in gene copy numbers assessed by uniplex or duplex ddPCR assays regarding CNV in the pfgch1 gene (p = 0.276). The accuracy and %RSD value of the duplex ddPCR assay were 95% and 4%, respectively, regarding pfgch1 GCN. In the P. falciparum field samples, pfmdr1 and pfplasmepsin2 GCNs were amplified in 15% and 27% of samples from Ubon Ratchathani, Thailand, while pfgch1 GCN was amplified in 50% of samples from Yala, Thailand. There was 100% agreement between the GCN results obtained from the ddPCR and qPCR assays (κ = 1.00). The results suggested that multiplex ddPCR assay is the optional assay for the accurate detection of gene copy number without requiring calibration standards, while the cost and required time are reduced. Based on the results of this study, criteria for GCN detection by ddPCR analysis were generated.
Conclusions: The developed ddPCR assays are simple, accurate, precise and cost-effective tools for detection of the CNVs in the pfmdr1, pfplasmepsin2 and pfgch1 genes of P. falciparum. The ddPCR assay is a useful additional tool for the surveillance of anti-malarial drug resistance.
Keywords: Plasmodium falciparum; ddPCR; pfgch1; pfmdr1; pfplasmepsin2.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
Antimalarial resistance risk in Mozambique detected by a novel quadruplex droplet digital PCR assay.Antimicrob Agents Chemother. 2024 Jul 9;68(7):e0034624. doi: 10.1128/aac.00346-24. Epub 2024 May 21. Antimicrob Agents Chemother. 2024. PMID: 38771031 Free PMC article.
-
Molecular characterization of Plasmodium falciparum antifolate resistance markers in Thailand between 2008 and 2016.Malar J. 2020 Mar 4;19(1):107. doi: 10.1186/s12936-020-03176-x. Malar J. 2020. PMID: 32127009 Free PMC article.
-
Low prevalence of copy number variation in pfmdr1 and pfpm2 in Plasmodium falciparum isolates from southern Angola.Malar J. 2025 Jan 10;24(1):5. doi: 10.1186/s12936-024-05240-2. Malar J. 2025. PMID: 39794826 Free PMC article.
-
Molecular surveillance for drug-resistant Plasmodium falciparum in clinical and subclinical populations from three border regions of Burma/Myanmar: cross-sectional data and a systematic review of resistance studies.Malar J. 2012 Sep 19;11:333. doi: 10.1186/1475-2875-11-333. Malar J. 2012. PMID: 22992214 Free PMC article.
-
Exploration of copy number variation in genes related to anti-malarial drug resistance in Plasmodium falciparum.Gene. 2020 Apr 30;736:144414. doi: 10.1016/j.gene.2020.144414. Epub 2020 Jan 30. Gene. 2020. PMID: 32006594 Review.
Cited by
-
High-throughput genotyping of Plasmodium vivax in the Peruvian Amazon via molecular inversion probes.Nat Commun. 2024 Nov 25;15(1):10219. doi: 10.1038/s41467-024-54731-y. Nat Commun. 2024. PMID: 39587110 Free PMC article.
-
Molecular tools are crucial for malaria elimination.Mol Biol Rep. 2024 Apr 20;51(1):555. doi: 10.1007/s11033-024-09496-4. Mol Biol Rep. 2024. PMID: 38642192 Review.
-
Antimalarial resistance risk in Mozambique detected by a novel quadruplex droplet digital PCR assay.Antimicrob Agents Chemother. 2024 Jul 9;68(7):e0034624. doi: 10.1128/aac.00346-24. Epub 2024 May 21. Antimicrob Agents Chemother. 2024. PMID: 38771031 Free PMC article.
-
Present and Future Applications of Digital PCR in Infectious Diseases Diagnosis.Diagnostics (Basel). 2024 Apr 29;14(9):931. doi: 10.3390/diagnostics14090931. Diagnostics (Basel). 2024. PMID: 38732345 Free PMC article. Review.
-
Advancing artemisinin resistance monitoring using a high sensitivity ddPCR assay for Pfkelch13 mutation detection in Asia.Sci Rep. 2025 Feb 10;15(1):4869. doi: 10.1038/s41598-025-86630-7. Sci Rep. 2025. PMID: 39929914 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

