Transcriptional activity of transposable elements along an elevational gradient in Arabidopsis arenosa
- PMID: 33639991
- PMCID: PMC7916287
- DOI: 10.1186/s13100-021-00236-0
Transcriptional activity of transposable elements along an elevational gradient in Arabidopsis arenosa
Abstract
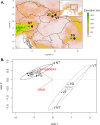
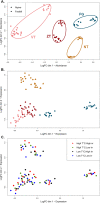
Background: Plant genomes can respond rapidly to environmental changes and transposable elements (TEs) arise as important drivers contributing to genome dynamics. Although some elements were reported to be induced by various abiotic or biotic factors, there is a lack of general understanding on how environment influences the activity and diversity of TEs. Here, we combined common garden experiment with short-read sequencing to investigate genomic abundance and expression of 2245 consensus TE sequences (containing retrotransposons and DNA transposons) in an alpine environment in Arabidopsis arenosa. To disentangle general trends from local differentiation, we leveraged four foothill-alpine population pairs from different mountain regions. Seeds of each of the eight populations were raised under four treatments that differed in temperature and irradiance, two factors varying with elevation. RNA-seq analysis was performed on leaves of young plants to test for the effect of elevation and subsequently of temperature and irradiance on expression of TE sequences.
Results: Genomic abundance of the 2245 consensus TE sequences varied greatly between the mountain regions in line with neutral divergence among the regions, representing distinct genetic lineages of A. arenosa. Accounting for intraspecific variation in abundance, we found consistent transcriptomic response for some TE sequences across the different pairs of foothill-alpine populations suggesting parallelism in TE expression. In particular expression of retrotransposon LTR Copia (e.g. Ivana and Ale clades) and LTR Gypsy (e.g. Athila and CRM clades) but also non-LTR LINE or DNA transposon TIR MuDR consistently varied with elevation of origin. TE sequences responding specifically to temperature and irradiance belonged to the same classes as well as additional TE clades containing potentially stress-responsive elements (e.g. LTR Copia Sire and Tar, LTR Gypsy Reina).
Conclusions: Our study demonstrated that the A. arenosa genome harbours a considerable diversity of TE sequences whose abundance and expression response varies across its native range. Some TE clades may contain transcriptionally active elements responding to a natural environmental gradient. This may further contribute to genetic variation between populations and may ultimately provide new regulatory mechanisms to face environmental challenges.
Keywords: Alpine environment; Arabidopsis arenosa; Common garden experiment; Parallelism; RNA-seq; Transposable elements.
Conflict of interest statement
The authors declare that have no competing interests.
Figures

Similar articles
-
Parallelism in gene expression between foothill and alpine ecotypes in Arabidopsis arenosa.Plant J. 2021 Mar;105(5):1211-1224. doi: 10.1111/tpj.15105. Epub 2021 Jan 4. Plant J. 2021. PMID: 33258160
-
Recent and dynamic transposable elements contribute to genomic divergence under asexuality.BMC Genomics. 2016 Nov 7;17(1):884. doi: 10.1186/s12864-016-3234-9. BMC Genomics. 2016. PMID: 27821059 Free PMC article.
-
Transposable element discovery and characterization of LTR-retrotransposon evolutionary lineages in the tropical fruit species Passiflora edulis.Mol Biol Rep. 2019 Dec;46(6):6117-6133. doi: 10.1007/s11033-019-05047-4. Epub 2019 Sep 24. Mol Biol Rep. 2019. PMID: 31549373
-
Human transposable elements in Repbase: genomic footprints from fish to humans.Mob DNA. 2018 Jan 4;9:2. doi: 10.1186/s13100-017-0107-y. eCollection 2018. Mob DNA. 2018. PMID: 29308093 Free PMC article. Review.
-
LTR-retrotransposons in plants: Engines of evolution.Gene. 2017 Aug 30;626:14-25. doi: 10.1016/j.gene.2017.04.051. Epub 2017 May 2. Gene. 2017. PMID: 28476688 Review.
Cited by
-
How to survive in the world's third poplar: Insights from the genome of the highest altitude woody plant, Hippophae tibetana (Elaeagnaceae).Front Plant Sci. 2022 Dec 14;13:1051587. doi: 10.3389/fpls.2022.1051587. eCollection 2022. Front Plant Sci. 2022. PMID: 36589082 Free PMC article.
-
Genome-wide analysis of horizontal transfer in non-model wild species from a natural ecosystem reveals new insights into genetic exchange in plants.PLoS Genet. 2023 Oct 19;19(10):e1010964. doi: 10.1371/journal.pgen.1010964. eCollection 2023 Oct. PLoS Genet. 2023. PMID: 37856455 Free PMC article.
-
Substrate specificity and protein stability drive the divergence of plant-specific DNA methyltransferases.Sci Adv. 2024 Nov 8;10(45):eadr2222. doi: 10.1126/sciadv.adr2222. Epub 2024 Nov 6. Sci Adv. 2024. PMID: 39504374 Free PMC article.
-
Genome-Wide Comparison of Structural Variations and Transposon Alterations in Soybean Cultivars Induced by Spaceflight.Int J Mol Sci. 2022 Nov 8;23(22):13721. doi: 10.3390/ijms232213721. Int J Mol Sci. 2022. PMID: 36430198 Free PMC article.
-
Transposable elements maintain genome-wide heterozygosity in inbred populations.Nat Commun. 2022 Nov 17;13(1):7022. doi: 10.1038/s41467-022-34795-4. Nat Commun. 2022. PMID: 36396660 Free PMC article.
References
-
- Bui QT, Grandbastien M-A. LTR retrotransposons as controlling elements of genome response to stress? Plant transposable elements. Springer. 2012. pp. 273–296.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

