COVID-19: Immunology, Immunopathogenesis and Potential Therapies
- PMID: 33641587
- PMCID: PMC7919479
- DOI: 10.1080/08830185.2021.1883600
COVID-19: Immunology, Immunopathogenesis and Potential Therapies
Abstract
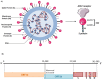
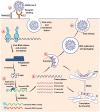
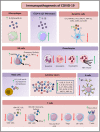
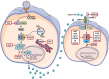
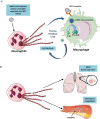
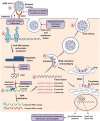
The Coronavirus Disease-2019 (COVID-19) imposed public health emergency and affected millions of people around the globe. As of January 2021, 100 million confirmed cases of COVID-19 along with more than 2 million deaths were reported worldwide. SARS-CoV-2 infection causes excessive production of pro-inflammatory cytokines thereby leading to the development of "Cytokine Storm Syndrome." This condition results in uncontrollable inflammation that further imposes multiple-organ-failure eventually leading to death. SARS-CoV-2 induces unrestrained innate immune response and impairs adaptive immune responses thereby causing tissue damage. Thus, understanding the foremost features and evolution of innate and adaptive immunity to SARS-CoV-2 is crucial in anticipating COVID-19 outcomes and in developing effective strategies to control the viral spread. In the present review, we exhaustively discuss the sequential key immunological events that occur during SARS-CoV-2 infection and are involved in the immunopathogenesis of COVID-19. In addition to this, we also highlight various therapeutic options already in use such as immunosuppressive drugs, plasma therapy and intravenous immunoglobulins along with various novel potent therapeutic options that should be considered in managing COVID-19 infection such as traditional medicines and probiotics.
Keywords: COVID-19; SARS-CoV-2; cytokine storm; immunology; immunotherapy.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
