Advanced molecular characterization of enteropathogenic Escherichia coli isolated from diarrheic camel neonates in Egypt
- PMID: 33642790
- PMCID: PMC7896916
- DOI: 10.14202/vetworld.2021.85-91
Advanced molecular characterization of enteropathogenic Escherichia coli isolated from diarrheic camel neonates in Egypt
Abstract
Background and aim: Camels are important livestock in Egypt on cultural and economic bases, but studies of etiological agents of camelid diseases are limited. The enteropathogen Escherichia coli is a cause of broad spectrum gastrointestinal infections among humans and animals, especially in developing countries. Severe infections can lead to death. The current study aimed to identify pathogenic E. coli strains that cause diarrhea in camel calves and characterize their virulence and drug resistance at a molecular level.
Materials and methods: Seventy fecal samples were collected from diarrheic neonatal camel calves in Giza Governorate during 2018-2019. Samples were cultured on a selective medium for E. coli, and positive colonies were confirmed biochemically, serotyped, and tested for antibiotic susceptibility. E. coli isolates were further confirmed through detection of the housekeeping gene, yaiO, and examined for the presence of virulence genes; traT and fimH and for genes responsible for antibiotic resistance, ampC, aadB, and mphA. The isolates in the important isolated serotype, E. coli O26, were examined for toxigenic genes and sequenced.
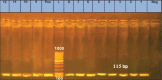
Results: The bacteriological and biochemical examination identified 12 E. coli isolates from 70 fecal samples (17.1%). Serotyping of these isolates showed four types: O26, four isolates, 33.3%; O103, O111, three isolates each, 25%; and O45, two isolates, 16.7%. The isolates showed resistance to vancomycin (75%) and ampicillin (66.6%), but were highly susceptible to ciprofloxacin, norfloxacin, and tetracycline (100%). The structural gene, yaiO (115 bp), was amplified from all 12 E. coli isolates and traT and fimH genes were amplified from 10 and 8 isolates, respectively. Antibiotic resistance genes, ampC, mphA, and aadB, were harbored in 9 (75%), 8 (66.6%), and 5 (41.7%), respectively. Seven isolates (58.3%) were MDR. Real-time-polymerase chain reaction of the O26 isolates identified one isolate harboring vt1, two with vt2, and one isolate with neither gene. Sequencing of the isolates revealed similarities to E. coli O157 strains.
Conclusion: Camels and other livestock suffer various diseases, including diarrhea often caused by microbial pathogens. Enteropathogenic E. coli serotypes were isolated from diarrheic neonatal camel calves. These isolates exhibited virulence and multiple drug resistance genes.
Keywords: camel; multidrug-resistant Escherichia coli; real-time polymerase chain reaction; sequencing; virulence.
Copyright: © Shahein, et al.
Figures


References
-
- Osman K.M, Mustafa A.M, Elhariri M, El-Hamed G.S.A. The distribution of Escherichia coli serovars, virulence genes, gene association and combinations and virulence genes encoding serotypes in pathogenic E. coli recovered from diarrheic calves, sheep and goat. Transbound. Emerg. Dis. 2013;60(1):69–78. - PubMed
-
- Galal H.M, Hakim A.S, Dorgham M.S. Phenotypic and virulence genes screening of Escherichia coli strains isolated from different sources in delta Egypt. Life Sci. J. 2013;10(2):352–361.
LinkOut - more resources
Full Text Sources
Other Literature Sources
