Early Diffusion of SARS-CoV-2 Infection in the Inner Area of the Italian Sardinia Island
- PMID: 33643227
- PMCID: PMC7907441
- DOI: 10.3389/fmicb.2020.628194
Early Diffusion of SARS-CoV-2 Infection in the Inner Area of the Italian Sardinia Island
Abstract
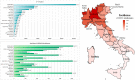
Background: Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has been responsible for the coronavirus disease 2019 (COVID-19) pandemic, which started as a severe pneumonia outbreak in Wuhan, China, in December 2019. Italy has been the first European country affected by the pandemic, registering a total of 300,363 cases and 35,741 deaths until September 24, 2020. The geographical distribution of SARS-CoV-2 in Italy during early 2020 has not been homogeneous, including regions severely affected as well as administrative areas being only slightly interested by the infection. Among the latter, Sardinia represents one of the lowest incidence areas likely due to its insular nature.
Methods: Next-generation sequencing of a small number of complete viral genomes from clinical samples and their virologic and phylogenetic characterization was performed.
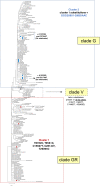
Results: We provide a first overview of the SARS-CoV-2 genomic diversity in Sardinia in the early phase of the March-May 2020 pandemic based on viral genomes isolated in the most inner regional hospital of the island. Our analysis revealed a remarkable genetic diversity in local SARS-CoV-2 viral genomes, showing the presence of at least four different clusters that can be distinguished by specific amino acid substitutions. Based on epidemiological information, these sequences can be linked to at least eight different clusters of infection, four of which likely originates from imported cases. In addition, the presence of amino acid substitutions that were not previously reported in Italian patients has been observed, asking for further investigations in a wider population to assess their prevalence and dynamics of emergence during the pandemic.
Conclusion: The present study provides a snapshot of the initial phases of the SARS-CoV-2 infection in inner area of the Sardinia Island, showing an unexpected genomic diversity.
Keywords: COVID-19; SARS-CoV-2; Sardinia Island; epidemiology; genome sequencing; molecular characterization; pandemic; phylogeny.
Copyright © 2021 Piras, Grandi, Monne, Asproni, Fancello, Fiamma, Mameli, Casu, lo Maglio, Palmas and Tramontano.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

