The Mitogenomes of Ophiostoma minus and Ophiostoma piliferum and Comparisons With Other Members of the Ophiostomatales
- PMID: 33643245
- PMCID: PMC7902536
- DOI: 10.3389/fmicb.2021.618649
The Mitogenomes of Ophiostoma minus and Ophiostoma piliferum and Comparisons With Other Members of the Ophiostomatales
Abstract
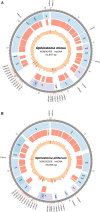
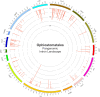
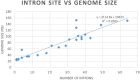
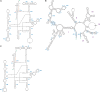
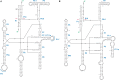
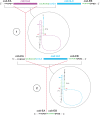
Fungi assigned to the Ophiostomatales are of economic concern as many are blue-stain fungi and some are plant pathogens. The mitogenomes of two blue-stain fungi, Ophiostoma minus and Ophiostoma piliferum, were sequenced and compared with currently available mitogenomes for other members of the Ophiostomatales. Species representing various genera within the Ophiostomatales have been examined for gene content, gene order, phylogenetic relationships, and the distribution of mobile elements. Gene synteny is conserved among the Ophiostomatales but some members were missing the atp9 gene. A genome wide intron landscape has been prepared to demonstrate the distribution of the mobile genetic elements (group I and II introns and homing endonucleases) and to provide insight into the evolutionary dynamics of introns among members of this group of fungi. Examples of complex introns or nested introns composed of two or three intron modules have been observed in some species. The size variation among the mitogenomes (from 23.7 kb to about 150 kb) is mostly due to the presence and absence of introns. Members of the genus Sporothrix sensu stricto appear to have the smallest mitogenomes due to loss of introns. The taxonomy of the Ophiostomatales has recently undergone considerable revisions; however, some lineages remain unresolved. The data showed that genera such as Raffaelea appear to be polyphyletic and the separation of Sporothrix sensu stricto from Ophiostoma is justified.
Keywords: Ophiostoma; blue stain fungi; complex introns; homing endonucleases; mitochondria; mobile introns.
Copyright © 2021 Zubaer, Wai, Patel, Perillo and Hausner.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Baidyaroy D., Hausner G., Hafez M., Michel F., Fulbright D. W., Bertrand H. (2011). A 971-bp insertion in the rns gene is associated with mitochondrial hypovirulence in a strain of Cryphonectria parasitica isolated from nature. Fungal Genet. Biol. 48 775–783. 10.1016/j.fgb.2011.05.006 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

