An Optimized High-Throughput Immuno-Plaque Assay for SARS-CoV-2
- PMID: 33643253
- PMCID: PMC7906992
- DOI: 10.3389/fmicb.2021.625136
An Optimized High-Throughput Immuno-Plaque Assay for SARS-CoV-2
Abstract
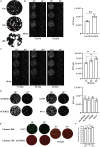
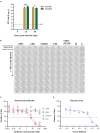
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) has been identified as the causative agent of coronavirus disease 2019 and is capable of human-to-human transmission and rapid global spread. The rapid emergence and global spread of SARS-CoV-2 has encouraged the establishment of a rapid, sensitive, and reliable viral detection and quantification methodology. Here, we present an alternative assay, termed immuno-plaque assay (iPA), which utilizes a combination of plaque assay and immunofluorescence techniques. We have extensively optimized the conditions for SARS-CoV-2 infection and demonstrated the great flexibility of iPA detection using several antibodies and dual-probing with two distinct epitope-specific antibodies. In addition, we showed that iPA could be utilized for ultra-high-throughput viral titration and neutralization assay within 24 h and is amenable to a 384-well format. These advantages will significantly accelerate SARS-CoV-2 research outcomes during this pandemic period.
Keywords: SARS-CoV-2; coronaviruses; immuno-plaque assay (iPA); viral quantification.
Copyright © 2021 Amarilla, Modhiran, Setoh, Peng, Sng, Liang, McMillan, Freney, Cheung, Chappell, Khromykh, Young and Watterson.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Agbulos D. S., Barelli L., Giordano B. V., Hunter F. F. (2016). Zika virus: quantification, propagation, detection, and storage. Curr. Protoc. Microbiol. 43 15D.4 1–15D.16. - PubMed
-
- Brien J. D., Lazear H. M., Diamond M. S. (2013). Propagation, quantification, detection, and storage of west nile virus. Curr. Protoc. Microbiol. 31 15D.3.1–15D.3.18. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

