Bioprospecting of Novel Extremozymes From Prokaryotes-The Advent of Culture-Independent Methods
- PMID: 33643258
- PMCID: PMC7902512
- DOI: 10.3389/fmicb.2021.630013
Bioprospecting of Novel Extremozymes From Prokaryotes-The Advent of Culture-Independent Methods
Abstract
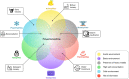
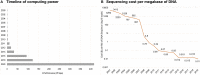
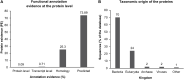
Extremophiles are remarkable organisms that thrive in the harshest environments on Earth, such as hydrothermal vents, hypersaline lakes and pools, alkaline soda lakes, deserts, cold oceans, and volcanic areas. These organisms have developed several strategies to overcome environmental stress and nutrient limitations. Thus, they are among the best model organisms to study adaptive mechanisms that lead to stress tolerance. Genetic and structural information derived from extremophiles and extremozymes can be used for bioengineering other nontolerant enzymes. Furthermore, extremophiles can be a valuable resource for novel biotechnological and biomedical products due to their biosynthetic properties. However, understanding life under extreme conditions is challenging due to the difficulties of in vitro cultivation and observation since > 99% of organisms cannot be cultivated. Consequently, only a minor percentage of the potential extremophiles on Earth have been discovered and characterized. Herein, we present a review of culture-independent methods, sequence-based metagenomics (SBM), and single amplified genomes (SAGs) for studying enzymes from extremophiles, with a focus on prokaryotic (archaea and bacteria) microorganisms. Additionally, we provide a comprehensive list of extremozymes discovered via metagenomics and SAGs.
Keywords: SAG; culture-independent methods; extremophile; halophiles; metagenomics; psychrophile; thermophiles.
Copyright © 2021 Sysoev, Grötzinger, Renn, Eppinger, Rueping and Karan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

