Microbial Community Dynamics of Soybean (Glycine max) Is Affected by Cropping Sequence
- PMID: 33643263
- PMCID: PMC7904696
- DOI: 10.3389/fmicb.2021.632280
Microbial Community Dynamics of Soybean (Glycine max) Is Affected by Cropping Sequence
Abstract
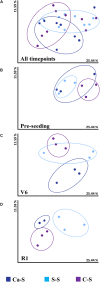
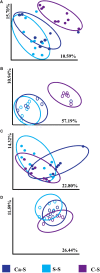
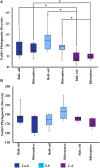
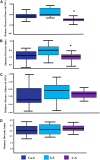
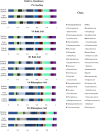
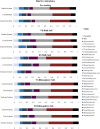
The microbial composition of the rhizosphere soil could be an important determinant of crop yield, pathogen resistance, and other beneficial attributes in plants. However, little is known about the impact of cropping sequences on microbial community dynamics, especially in economically important species like soybean. Using 2-year crop sequences of corn-soybean, canola-soybean, and soybean-soybean, we investigated how crops from the previous growing season influenced the structure of the microbiome in both the bulk soil and soybean rhizosphere. A combination of marker-based Illumina sequencing and bioinformatics analyses was used to show that bacterial species richness and evenness in the soybean rhizosphere soil were similar following canola and soybean compared to a previous corn sequence. However, fungal species richness and evenness remained unaffected by crop sequence. In addition, bacterial and fungal species diversity in both the bulk and soybean rhizosphere soil were not influenced by crop sequence. Lastly, the corn-soybean sequence significantly differed in the relative abundance of certain bacterial and fungal classes in both the soybean rhizosphere and bulk soil. While canola-soybean and a continuous soybean sequence did not, suggesting that a preceding corn sequence may reduce the occurrence of overall bacterial and fungal community members. For the present study, crop sequence impacts bacterial diversity and richness in both the bulk soil and soybean rhizosphere soil whereas fungal diversity and richness are resilient to crop sequence practices. Together, these findings could help drive decision making for annual crop and soil management practices.
Keywords: canola; corn; crop sequence; phytomicrobiome; soybean.
Copyright © 2021 Bolaji, Wan, Manchur, Lawley, de Kievit, Fernando and Belmonte.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Agomoh I. V., Drury C. F., Phillips L. A., Reynolds W. D., Yang X. (2020). Increasing crop diversity in wheat rotations increases yields but decreases soil health. Soil Sci. Soc. Am. J. 84 170–181. 10.1002/saj2.20000 - DOI
-
- Aguirre-von-Wobeser E., Rocha-Estrada J., Shapiro L. R., de la Torre M. (2018). Enrichment of Verrucomicrobia, Actinobacteria and Burkholderiales drives selection of bacterial community from soil by maize roots in a traditional milpa agroecosystem. PLoS One 13:e0208852. 10.1371/journal.pone.0208852 - DOI - PMC - PubMed
-
- Ai C., Zhang S., Zhang X., Guo D., Zhou W., Huang S. (2018). Distinct responses of soil bacterial and fungal communities to changes in fertilization regime and crop rotation. Geoderma 319 156–166. 10.1016/j.geoderma.2018.01.010 - DOI
-
- Ashworth A. J., DeBruyn J. M., Allen F. L., Radosevich M., Owens P. R. (2017). Microbial community structure is affected by cropping sequences and poultry litter under long-term no-tillage. Soil Biol. Biochem. 114 210–219. 10.1016/j.soilbio.2017.07.019 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

