Optimized Culture Conditions for Improved Growth and Functional Differentiation of Mouse and Human Colon Organoids
- PMID: 33643277
- PMCID: PMC7906999
- DOI: 10.3389/fimmu.2020.547102
Optimized Culture Conditions for Improved Growth and Functional Differentiation of Mouse and Human Colon Organoids
Abstract
Background & aims: Diligent side-by-side comparisons of how different methodologies affect growth efficiency and quality of intestinal colonoids have not been performed leaving a gap in our current knowledge. Here, we summarize our efforts to optimize culture conditions for improved growth and functional differentiation of mouse and human colon organoids.
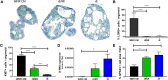
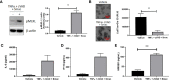
Methods: Mouse and human colon organoids were grown in four different media. Media-dependent long-term growth was measured by quantifying surviving organoids via imaging and a cell viability readout over five passages. The impact of diverse media on differentiation was assessed by quantifying the number of epithelial cell types using markers for enterocytes, stem cells, Goblet cells, and enteroendocrine cells by qPCR and histology upon removal of growth factors.
Results: In contrast to Wnt3a-conditioned media, media supplemented with recombinant Wnt3a alone did not support long-term survival of human or mouse colon organoids. Mechanistically, this observation can be attributed to the fact that recombinant Wnt3a did not support stem cell survival or proliferation as demonstrated by decreased LGR5 and Ki67 expression. When monitoring expression of markers for epithelial cell types, the highest level of organoid differentiation was observed after combined removal of Wnt3a, Noggin, and R-spondin from Wnta3a-conditioned media cultures.
Conclusion: Our study defined Wnt3a-containing conditioned media as optimal for growth and survival of human and mouse organoids. Furthermore, we established that the combined removal of Wnt3a, Noggin, and R-spondin results in optimal differentiation. This study provides a step forward in optimizing conditions for intestinal organoid growth to improve standardization and reproducibility of this model platform.
Keywords: Goblet cells; Wnt3 signaling; adult stem cells; colonoids; differentiation; enteroids; intestinal epithelial cells; intestinal organoids.
Copyright © 2021 Wilson, Mayo, Melim, Knight, Patnaude, Wu, Phillips, Westmoreland, Dunstan, Fiebiger and Terrillon.
Conflict of interest statement
All authors are employees of AbbVie. The design, study conduct, and financial support for this research were provided by AbbVie. AbbVie participated in the interpretation of data, review, and approval of the publication.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

