The Peripheral Blood Transcriptome Is Correlated With PET Measures of Lung Inflammation During Successful Tuberculosis Treatment
- PMID: 33643286
- PMCID: PMC7902901
- DOI: 10.3389/fimmu.2020.596173
The Peripheral Blood Transcriptome Is Correlated With PET Measures of Lung Inflammation During Successful Tuberculosis Treatment
Abstract
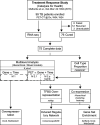
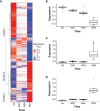
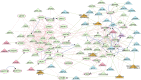
Pulmonary tuberculosis (PTB) is characterized by lung granulomas, inflammation and tissue destruction. Here we used within-subject peripheral blood gene expression over time to correlate with the within-subject lung metabolic activity, as measured by positron emission tomography (PET) to identify biological processes and pathways underlying overall resolution of lung inflammation. We used next-generation RNA sequencing and [18F]FDG PET-CT data, collected at diagnosis, week 4, and week 24, from 75 successfully cured PTB patients, with the [18F]FDG activity as a surrogate for lung inflammation. Our linear mixed-effects models required that for each individual the slope of the line of [18F]FDG data in the outcome and the slope of the peripheral blood transcript expression data correlate, i.e., the slopes of the outcome and explanatory variables had to be similar. Of 10,295 genes that changed as a function of time, we identified 639 genes whose expression profiles correlated with decreasing [18F]FDG uptake levels in the lungs. Gene enrichment over-representation analysis revealed that numerous biological processes were significantly enriched in the 639 genes, including several well known in TB transcriptomics such as platelet degranulation and response to interferon gamma, thus validating our novel approach. Others not previously associated with TB pathobiology included smooth muscle contraction, a set of pathways related to mitochondrial function and cell death, as well as a set of pathways connecting transcription, translation and vesicle formation. We observed up-regulation in genes associated with B cells, and down-regulation in genes associated with platelet activation. We found 254 transcription factor binding sites to be enriched among the 639 gene promoters. In conclusion, we demonstrated that of the 10,295 gene expression changes in peripheral blood, only a subset of 639 genes correlated with inflammation in the lungs, and the enriched pathways provide a description of the biology of resolution of lung inflammation as detectable in peripheral blood. Surprisingly, resolution of PTB inflammation is positively correlated with smooth muscle contraction and, extending our previous observation on mitochondrial genes, shows the presence of mitochondrial stress. We focused on pathway analysis which can enable therapeutic target discovery and potential modulation of the host response to TB.
Keywords: RNA-sequencing; [18F]FDG PET-CT; gene expression; mixed-effect models; pathway analysis; transcription factor binding site; treatment response; tuberculosis.
Copyright © 2021 Odia, Malherbe, Meier, Maasdorp, Kleynhans, du Plessis, Loxton, Zak, Thompson, Duffy, Kuivaniemi, Ronacher, Winter, Walzl, Tromp and the Catalysis TB-Biomarker Consortium.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- World Health Organization Global tuberculosis report 2019. (2019). p. 283.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

