Multipathogen Analysis of IgA and IgG Antigen Specificity for Selected Pathogens in Milk Produced by Women From Diverse Geographical Regions: The INSPIRE Study
- PMID: 33643297
- PMCID: PMC7905217
- DOI: 10.3389/fimmu.2020.614372
Multipathogen Analysis of IgA and IgG Antigen Specificity for Selected Pathogens in Milk Produced by Women From Diverse Geographical Regions: The INSPIRE Study
Abstract
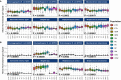
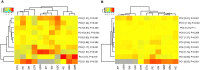
Breastfeeding provides defense against infectious disease during early life. The mechanisms underlying this protection are complex but likely include the vast array of immune cells and components, such as immunoglobulins, in milk. Simply characterizing the concentrations of these bioactives, however, provides only limited information regarding their potential relationships with disease risk in the recipient infant. Rather, understanding pathogen and antigen specificity profiles of milk-borne immunoglobulins might lead to a more complete understanding of how maternal immunity impacts infant health and wellbeing. Milk produced by women living in 11 geographically dispersed populations was applied to a protein microarray containing antigens from 16 pathogens, including diarrheagenic E. coli, Shigella spp., Salmonella enterica serovar Typhi, Staphylococcus aureus, Streptococcus pneumoniae, Mycobacterium tuberculosis and other pathogens of global health concern, and specific IgA and IgG binding was measured. Our analysis identified novel disease-specific antigen responses and suggests that some IgA and IgG responses vary substantially within and among populations. Patterns of antibody reactivity analyzed by principal component analysis and differential reactivity analysis were associated with either lower-to-middle-income countries (LMICs) or high-income countries (HICs). Antibody levels were generally higher in LMICs than HICs, particularly for Shigella and diarrheagenic E. coli antigens, although sets of S. aureus, S. pneumoniae, and some M. tuberculosis antigens were more reactive in HICs. Differential responses were typically specific to canonical immunodominant antigens, but a set of nondifferential but highly reactive antibodies were specific to antigens possibly universally recognized by antibodies in human milk. This approach provides a promising means to understand how breastfeeding and human milk protect (or do not protect) infants from environmentally relevant pathogens. Furthermore, this approach might lead to interventions to boost population-specific immunity in at-risk breastfeeding mothers and their infants.
Keywords: IgA; IgG; breastfeeding; breastmilk; human milk; immunoglobulins; pathogen; protein array.
Copyright © 2021 McGuire, Randall, Seppo, Järvinen, Meehan, Gindola, Williams, Sellen, Kamau-Mbuthia, Kamundia, Mbugua, Moore, Prentice, Foster, Otoo, Rodríguez, Pareja, Bode, McGuire and Campo.
Conflict of interest statement
AR and JC are employees of Antigen Discovery Incorporated, a company that carries patents related to the protein array analyses used here and the location where the arrays were conducted. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- World Health Organization Exclusive breastfeeding for six months best for babies everywhere. Statement. WHO; (2011) Available at: http://www.who.int/mediacentre/news/statements/2011/breastfeeding_201101..., January 15, 2011.
-
- Oddy WH. Infant feeding and obesity risk in the child. Breastfeed Rev (2012) 20(2):7–12. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

