Comparative Analyses of Euonymus Chloroplast Genomes: Genetic Structure, Screening for Loci With Suitable Polymorphism, Positive Selection Genes, and Phylogenetic Relationships Within Celastrineae
- PMID: 33643327
- PMCID: PMC7905392
- DOI: 10.3389/fpls.2020.593984
Comparative Analyses of Euonymus Chloroplast Genomes: Genetic Structure, Screening for Loci With Suitable Polymorphism, Positive Selection Genes, and Phylogenetic Relationships Within Celastrineae
Abstract
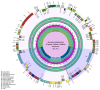
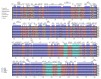
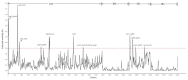
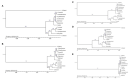
In this study, we assembled and annotated the chloroplast (cp) genome of the Euonymus species Euonymus fortunei, Euonymus phellomanus, and Euonymus maackii, and performed a series of analyses to investigate gene structure, GC content, sequence alignment, and nucleic acid diversity, with the objectives of identifying positive selection genes and understanding evolutionary relationships. The results indicated that the Euonymus cp genome was 156,860-157,611bp in length and exhibited a typical circular tetrad structure. Similar to the majority of angiosperm chloroplast genomes, the results yielded a large single-copy region (LSC) (85,826-86,299bp) and a small single-copy region (SSC) (18,319-18,536bp), separated by a pair of sequences (IRA and IRB; 26,341-26,700bp) with the same encoding but in opposite directions. The chloroplast genome was annotated to 130-131 genes, including 85-86 protein coding genes, 37 tRNA genes, and eight rRNA genes, with GC contents of 37.26-37.31%. The GC content was variable among regions and was highest in the inverted repeat (IR) region. The IR boundary of Euonymus happened expanding resulting that the rps19 entered into IR region and doubled completely. Such fluctuations at the border positions might be helpful in determining evolutionary relationships among Euonymus. The simple-sequence repeats (SSRs) of Euonymus species were composed primarily of single nucleotides (A)n and (T)n, and were mostly 10-12bp in length, with an obvious A/T bias. We identified several loci with suitable polymorphism with the potential use as molecular markers for inferring the phylogeny within the genus Euonymus. Signatures of positive selection were seen in rpoB protein encoding genes. Based on data from the whole chloroplast genome, common single copy genes, and the LSC, SSC, and IR regions, we constructed an evolutionary tree of Euonymus and related species, the results of which were consistent with traditional taxonomic classifications. It showed that E. fortunei sister to the Euonymus japonicus, whereby E. maackii appeared as sister to Euonymus hamiltonianus. Our study provides important genetic information to support further investigations into the phylogenetic development and adaptive evolution of Euonymus species.
Keywords: Euonymus; adaptive evolution; chloroplast genome; molecular marker; phylograms.
Copyright © 2021 Li, Dong, Liu, Yu, Yang and Huang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Abdullah, Henriquez C. L., Mehmood F., Carlsen M. M., Islam M., Waheed M. T., et al. (2020a). Complete chloroplast genomes of Anthurium huixtlense and Pothos scandens (Pothoideae, Araceae): unique inverted repeat expansion and contraction affect rate of evolution. J. Mol. Evol. 88, 562–574. 10.1007/s00239-020-09958-w, PMID: - DOI - PMC - PubMed
-
- Abdullah, Mehmood F., Shahzadi I., Ali Z., Islam M., Naeem M., et al. (2020c). Correlations among oligonucleotide repeats, nucleotide substitutions, and insertion–deletion mutations in chloroplast genomes of plant family Malvaceae. J. Syst. Evol. 1–15. 10.1111/jse.12585 - DOI
-
- Abdullah, Shahzadi I., Mehmood F., Ali Z., Malik M. S., Waseem S., et al. (2019a). Comparative analyses of chloroplast genomes among three Firmiana species: identification of mutational hotspots and phylogenetic relationship with other species of Malvaceae. Plant Gene 19:100199. 10.1016/j.plgene.2019.100199 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

