Abiotic Stress-Responsive miRNA and Transcription Factor-Mediated Gene Regulatory Network in Oryza sativa: Construction and Structural Measure Study
- PMID: 33643383
- PMCID: PMC7907651
- DOI: 10.3389/fgene.2021.618089
Abiotic Stress-Responsive miRNA and Transcription Factor-Mediated Gene Regulatory Network in Oryza sativa: Construction and Structural Measure Study
Abstract
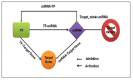
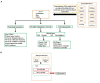
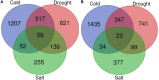
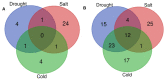
Climate changes and environmental stresses have a consequential association with crop plant growth and yield, meaning it is necessary to cultivate crops that have tolerance toward the changing climate and environmental disturbances such as water stress, temperature fluctuation, and salt toxicity. Recent studies have shown that trans-acting regulatory elements, including microRNAs (miRNAs) and transcription factors (TFs), are emerging as promising tools for engineering naive improved crop varieties with tolerance for multiple environmental stresses and enhanced quality as well as yield. However, the interwoven complex regulatory function of TFs and miRNAs at transcriptional and post-transcriptional levels is unexplored in Oryza sativa. To this end, we have constructed a multiple abiotic stress responsive TF-miRNA-gene regulatory network for O. sativa using a transcriptome and degradome sequencing data meta-analysis approach. The theoretical network approach has shown the networks to be dense, scale-free, and small-world, which makes the network stable. They are also invariant to scale change where an efficient, quick transmission of biological signals occurs within the network on extrinsic hindrance. The analysis also deciphered the existence of communities (cluster of TF, miRNA, and genes) working together to help plants in acclimatizing to multiple stresses. It highlighted that genes, TFs, and miRNAs shared by multiple stress conditions that work as hubs or bottlenecks for signal propagation, for example, during the interaction between stress-responsive genes (TFs/miRNAs/other genes) and genes involved in floral development pathways under multiple environmental stresses. This study further highlights how the fine-tuning feedback mechanism works for balancing stress tolerance and how timely flowering enable crops to survive in adverse conditions. This study developed the abiotic stress-responsive regulatory network, APRegNet database (http://lms.snu.edu.in/APRegNet), which may help researchers studying the roles of miRNAs and TFs. Furthermore, it advances current understanding of multiple abiotic stress tolerance mechanisms.
Keywords: Oryza sativa; microRNA; post-transcriptional regulation; regulatory network; target mimics; transcription factor.
Copyright © 2021 Sharma, Upadhyay, Bhattacharya and Singh.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Barabási A.-L., Albert R. (1999). Emergence of scaling in random networks. Science 308 639–641. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

