Covalent conjugation of extracellular vesicles with peptides and nanobodies for targeted therapeutic delivery
- PMID: 33643546
- PMCID: PMC7886705
- DOI: 10.1002/jev2.12057
Covalent conjugation of extracellular vesicles with peptides and nanobodies for targeted therapeutic delivery
Abstract
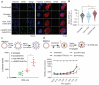
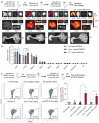
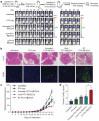
Natural extracellular vesicles (EVs) are ideal drug carriers due to their remarkable biocompatibility. Their delivery specificity can be achieved by the conjugation of targeting ligands. However, existing methods to engineer target-specific EVs are tedious or inefficient, having to compromise between harsh chemical treatments and transient interactions. Here, we describe a novel method for the covalent conjugation of EVs with high copy numbers of targeting moieties using protein ligases. Conjugation of EVs with either an epidermal growth factor receptor (EGFR)-targeting peptide or anti-EGFR nanobody facilitates their accumulation in EGFR-positive cancer cells, both in vitro and in vivo. Systemic delivery of paclitaxel by EGFR-targeting EVs at a low dose significantly increases drug efficacy in a xenografted mouse model of EGFR-positive lung cancer. The method is also applicable to the conjugation of EVs with peptides and nanobodies targeting other receptors, such as HER2 and SIRP alpha, and the conjugated EVs can deliver RNA in addition to small molecules, supporting the versatile application of EVs in cancer therapies. This simple, yet efficient and versatile method for the stable surface modification of EVs bypasses the need for genetic and chemical modifications, thus facilitating safe and specific delivery of therapeutic payloads to target cells.
Keywords: conjugation; delivery; extracellular vesicles; targeted; therapeutics.
© 2021 The Authors. Journal of Extracellular Vesicles published by Wiley Periodicals, LLC on behalf of the International Society for Extracellular Vesicles.
Conflict of interest statement
Minh TN Le and Jiahai Shi are scientific co‐founders, and advisors of Carmine Therapeutics. Other authors declare no conflict of interest.
Figures

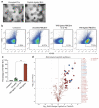
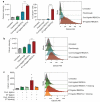
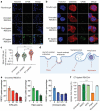
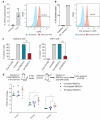

References
-
- Alvarez‐Erviti, L. , Seow, Y. , Yin, H. , Betts, C. , Lakhal, S. , & Wood, M. J. A. (2011). Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nature Biotechnology 29, 341–345. - PubMed
-
- Andaloussi, S. E. L. , Mäger, I. , Breakefield, X. O. , & Wood, M. J. A. (2013). Extracellular vesicles: Biology and emerging therapeutic opportunities. Nature Reviews Drug Discovery 12, 347–357. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

