Senescent Tumor Cells Build a Cytokine Shield in Colorectal Cancer
- PMID: 33643790
- PMCID: PMC7887594
- DOI: 10.1002/advs.202002497
Senescent Tumor Cells Build a Cytokine Shield in Colorectal Cancer
Abstract
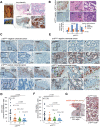
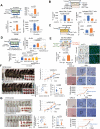
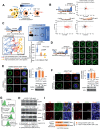
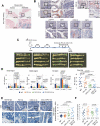
Cellular senescence can either support or inhibit cancer progression. Here, it is shown that intratumoral infiltration of CD8+ T cells is negatively associated with the proportion of senescent tumor cells in colorectal cancer (CRC). Gene expression analysis reveals increased expression of C-X-C motif chemokine ligand 12 (CXCL12) and colony stimulating factor 1 (CSF1) in senescent tumor cells. Senescent tumor cells inhibit CD8+ T cell infiltration by secreting a high concentration of CXCL12, which induces a loss of CXCR4 in T cells that result in impaired directional migration. CSF1 from senescent tumor cells enhance monocyte differentiation into M2 macrophages, which inhibit CD8+ T cell activation. Neutralization of CXCL12/CSF1 increases the effect of anti-PD1 antibody in allograft tumors. Furthermore, inhibition of CXCL12 from senescent tumor cells enhances T cell infiltration and results in reducing the number and size of tumors in azoxymethane (AOM)/dextran sulfate sodium (DSS)-induced CRC. These findings suggest senescent tumor cells generate a cytokine barrier protecting nonsenescent tumor cells from immune attack and provide a new target for overcoming the immunotherapy resistance of CRC.
Keywords: CD8+ T cells; CXCL12; cancer immunotherapy; colorectal cancers; senescent tumor cells.
© 2021 The Authors. Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Braig M., Lee S., Loddenkemper C., Rudolph C., Peters A. H., Schlegelberger B., Stein H., Dorken B., Jenuwein T., Schmitt C. A., Nature 2005, 436, 660. - PubMed
-
- Michaloglou C., Vredeveld L. C., Soengas M. S., Denoyelle C., Kuilman T., van der Horst C. M., Majoor D. M., Shay J. W., Mooi W. J., Peeper D. S., Nature 2005, 436, 720. - PubMed
-
- Hernandez‐Segura A., Nehme J., Demaria M., Trends Cell Biol. 2018, 28, 436. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
