Challenges Associated With the Formation of Recombinant Protein Inclusion Bodies in Escherichia coli and Strategies to Address Them for Industrial Applications
- PMID: 33644021
- PMCID: PMC7902521
- DOI: 10.3389/fbioe.2021.630551
Challenges Associated With the Formation of Recombinant Protein Inclusion Bodies in Escherichia coli and Strategies to Address Them for Industrial Applications
Abstract
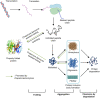
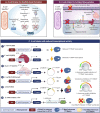
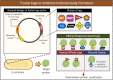
Recombinant proteins are becoming increasingly important for industrial applications, where Escherichia coli is the most widely used bacterial host for their production. However, the formation of inclusion bodies is a frequently encountered challenge for producing soluble and functional recombinant proteins. To overcome this hurdle, different strategies have been developed through adjusting growth conditions, engineering host strains of E. coli, altering expression vectors, and modifying the proteins of interest. These approaches will be comprehensively highlighted with some of the new developments in this review. Additionally, the unique features of protein inclusion bodies, the mechanism and influencing factors of their formation, and their potential advantages will also be discussed.
Keywords: E. coli; industrial applications; protein folding; protein inclusion bodies; recombinant proteins.
Copyright © 2021 Bhatwa, Wang, Hassan, Abraham, Li and Zhou.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

