FOntCell: Fusion of Ontologies of Cells
- PMID: 33644039
- PMCID: PMC7905052
- DOI: 10.3389/fcell.2021.562908
FOntCell: Fusion of Ontologies of Cells
Abstract
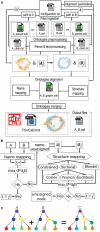
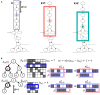
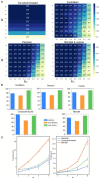
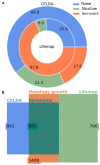
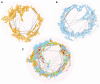
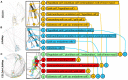
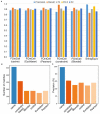
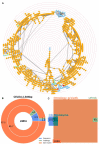
High-throughput cell-data technologies such as single-cell RNA-seq create a demand for algorithms for automatic cell classification and characterization. There exist several cell classification ontologies with complementary information. However, one needs to merge them to synergistically combine their information. The main difficulty in merging is to match the ontologies since they use different naming conventions. Therefore, we developed an algorithm that merges ontologies by integrating the name matching between class label names with the structure mapping between the ontology elements based on graph convolution. Since the structure mapping is a time consuming process, we designed two methods to perform the graph convolution: vectorial structure matching and constraint-based structure matching. To perform the vectorial structure matching, we designed a general method to calculate the similarities between vectors of different lengths for different metrics. Additionally, we adapted the slower Blondel method to work for structure matching. We implemented our algorithms into FOntCell, a software module in Python for efficient automatic parallel-computed merging/fusion of ontologies in the same or similar knowledge domains. FOntCell can unify dispersed knowledge from one domain into a unique ontology in OWL format and iteratively reuse it to continuously adapt ontologies with new data endlessly produced by data-driven classification methods, such as of the Human Cell Atlas. To navigate easily across the merged ontologies, it generates HTML files with tabulated and graphic summaries, and interactive circular Directed Acyclic Graphs. We used FOntCell to merge the CELDA, LifeMap and LungMAP Human Anatomy cell ontologies into a comprehensive cell ontology. We compared FOntCell with tools used for the alignment of mouse and human anatomy ontologies task proposed by the Ontology Alignment Evaluation Initiative (OAEI) and found that the Fβ alignment accuracies of FOntCell are above the geometric mean of the other tools; more importantly, it outperforms significantly the best OAEI tools in cell ontology alignment in terms of Fβ alignment accuracies.
Keywords: Human Cell Atlas (HCA); Ontology Alignment Evaluation Initiative (OAEI); automatic ontology merging; cell ontology; ontology alignment; ontology merging.
Copyright © 2021 Cabau-Laporta, Ascensión, Arrospide-Elgarresta, Gerovska and Araúzo-Bravo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The handling editor declared a past collaboration with one of the authors MA-B.
Figures
Similar articles
-
Matching biomedical ontologies with GCN-based feature propagation.Math Biosci Eng. 2022 Jun 9;19(8):8479-8504. doi: 10.3934/mbe.2022394. Math Biosci Eng. 2022. PMID: 35801474
-
Owlready: Ontology-oriented programming in Python with automatic classification and high level constructs for biomedical ontologies.Artif Intell Med. 2017 Jul;80:11-28. doi: 10.1016/j.artmed.2017.07.002. Epub 2017 Aug 14. Artif Intell Med. 2017. PMID: 28818520
-
Interactive biomedical ontology matching.PLoS One. 2019 Apr 17;14(4):e0215147. doi: 10.1371/journal.pone.0215147. eCollection 2019. PLoS One. 2019. PMID: 30995257 Free PMC article.
-
Experiences from the anatomy track in the ontology alignment evaluation initiative.J Biomed Semantics. 2017 Dec 4;8(1):56. doi: 10.1186/s13326-017-0166-5. J Biomed Semantics. 2017. PMID: 29202830 Free PMC article. Review.
-
Evaluation of ontology structural metrics based on public repository data.Brief Bioinform. 2020 Mar 23;21(2):473-485. doi: 10.1093/bib/bbz009. Brief Bioinform. 2020. PMID: 30715146 Review.
References
-
- Ascension A. M., Araúzo-Bravo M. J. (2020). “BigMPI4py: python module for parallelization of big data objects discloses germ layer specific DNA demethylation motifs,” in IEEE/ACM Transactions on Computing Biology and Bioinformatics (New York, NY: IEEE; ). - PubMed
-
- Blondel V. D., Gajardo A., Heymans M., Senellart P., Van Dooren P. (2004). A measure of similarity between graph vertices: applications to synonym extraction and web searching. SIAM Rev. 46, 647–666. 10.1137/S0036144502415960 - DOI
-
- Busse J., Humm B., Lübbert C., Moelter F., Reibold A., Rewald M., et al. (2015). Actually, what does “ontology” mean?: A term coined by philosophy in the light of different scientific disciplines. J. Comp. Inform. Technol. 23, 29–41. 10.2498/cit.1002508 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

