Molecular characterization of the stress network in individuals at risk for schizophrenia
- PMID: 33644266
- PMCID: PMC7893486
- DOI: 10.1016/j.ynstr.2021.100307
Molecular characterization of the stress network in individuals at risk for schizophrenia
Abstract
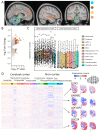
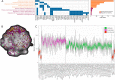
The biological mechanisms underlying inter-individual differences in human stress reactivity remain poorly understood. We aimed to identify the molecular underpinning of aberrant neural stress sensitivity in individuals at risk for schizophrenia. Linking mRNA expression data from the Allen Human Brain Atlas to task-based fMRI revealed 201 differentially expressed genes in cortex-specific brain regions differentially activated by stress in individuals with low (healthy siblings of schizophrenia patients) or high (healthy controls) stress sensitivity. These genes are associated with stress-related psychiatric disorders (e.g. schizophrenia and anxiety) and include markers for specific neuronal populations (e.g. ADCYAP1, GABRB1, SSTR1, and TNFRSF12A), neurotransmitter receptors (e.g. GRIN3A, SSTR1, GABRB1, and HTR1E), and signaling factors that interact with the corticosteroid receptor and hypothalamic-pituitary-adrenal axis (e.g. ADCYAP1, IGSF11, and PKIA). Overall, the identified genes potentially underlie altered stress reactivity in individuals at risk for schizophrenia and other psychiatric disorders and play a role in mounting an adaptive stress response in at-risk individuals, making them potentially druggable targets for stress-related diseases.
Keywords: Molecular correlates; Stress network; Stress reactivity; Stress sensitivity; Stress-related diseases.
© 2021 The Authors.
Figures


References
-
- Argyropoulos S.V., Landau S., Kalidindi S., Toulopoulou T., Castle D.J., Murray R.M., Picchioni M.M. Twins discordant for schizophrenia: psychopathology of the non-schizophrenic co-twins. Acta Psychiatr. Scand. 2008;118(3):214–219. - PubMed
-
- Baca-Garcia E., Vaquero-Lorenzo C., Perez-Rodriguez M.M., Gratacos M., Bayes M., Santiago-Mozos R., Leiva-Murillo J.M., de Prado-Cumplido M., Artes-Rodriguez A., Ceverino A., Diaz-Sastre C., Fernandez-Navarro P., Costas J., Fernandez-Piqueras J., Diaz-Hernandez M., de Leon J., Baca-Baldomero E., Saiz-Ruiz J., Mann J.J., Parsey R.V., Carracedo A., Estivill X., Oquendo M.A. Nucleotide variation in central nervous system genes among male suicide attempters. Am J Med Genet B Neuropsychiatr Genet. 2010;153B(1):208–213. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

