Reaction Pathway Sampling and Free-Energy Analyses for Multimeric Protein Complex Disassembly by Employing Hybrid Configuration Bias Monte Carlo/Molecular Dynamics Simulation
- PMID: 33644582
- PMCID: PMC7905796
- DOI: 10.1021/acsomega.0c05579
Reaction Pathway Sampling and Free-Energy Analyses for Multimeric Protein Complex Disassembly by Employing Hybrid Configuration Bias Monte Carlo/Molecular Dynamics Simulation
Abstract
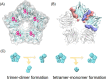
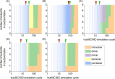
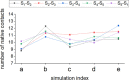
Physicochemical characterization of multimeric biomacromolecule assembly and disassembly processes is a milestone to understand the mechanisms for biological phenomena at the molecular level. Mass spectroscopy (MS) and structural bioinformatics (SB) approaches have become feasible to identify subcomplexes involved in assembly and disassembly, while they cannot provide atomic information sufficient for free-energy calculation to characterize transition mechanism between two different sets of subcomplexes. To combine observations derived from MS and SB approaches with conventional free-energy calculation protocols, we here designed a new reaction pathway sampling method by employing hybrid configuration bias Monte Carlo/molecular dynamics (hcbMC/MD) scheme and applied it to simulate the disassembly process of serum amyloid P component (SAP) pentamer. The results we obtained are consistent with those of the earlier MS and SB studies with respect to SAP subcomplex species and the initial stage of SAP disassembly processes. Furthermore, we observed a novel dissociation event, ring-opening reaction of SAP pentamer. Employing free-energy calculation combined with the hcbMC/MD reaction pathway trajectories, we moreover obtained experimentally testable observations on (1) reaction time of the ring-opening reaction and (2) importance of Asp42 and Lys117 for stable formation of SAP oligomer.
© 2021 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures





References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

