Single-cell analyses reveal YAP/TAZ as regulators of stemness and cell plasticity in Glioblastoma
- PMID: 33644767
- PMCID: PMC7116831
- DOI: 10.1038/s43018-020-00150-z
Single-cell analyses reveal YAP/TAZ as regulators of stemness and cell plasticity in Glioblastoma
Abstract
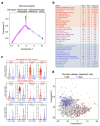
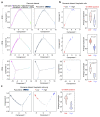
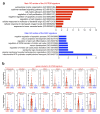
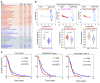
Glioblastoma (GBM) is a devastating human malignancy. GBM stem-like cells (GSCs) drive tumor initiation and progression. Yet, the molecular determinants defining GSCs in their native state in patients remain poorly understood. Here we used single cell datasets and identified GSCs at the apex of the differentiation hierarchy of GBM. By reconstructing the GSCs' regulatory network, we identified the YAP/TAZ coactivators as master regulators of this cell state, irrespectively of GBM subtypes. YAP/TAZ are required to install GSC properties in primary cells downstream of multiple oncogenic lesions, and required for tumor initiation and maintenance in vivo in different mouse and human GBM models. YAP/TAZ act as main roadblock of GSC differentiation and their inhibition irreversibly lock differentiated GBM cells into a non-tumorigenic state, preventing plasticity and regeneration of GSC-like cells. Thus, GSC identity is linked to a key molecular hub integrating genetics and microenvironmental inputs within the multifaceted biology of GBM.
Conflict of interest statement
Competing interests The authors declare no competing interests.
Figures

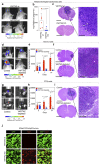
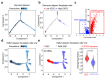

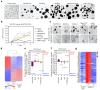
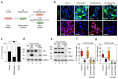

Comment in
-
Seeing the GBM diversity spectrum.Nat Cancer. 2021 Feb;2(2):135-137. doi: 10.1038/s43018-021-00176-x. Nat Cancer. 2021. PMID: 35122081 No abstract available.
References
-
- Singh SK, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. - PubMed
-
- Galli R, et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer research. 2004;64:7011–7021. - PubMed
-
- Bao S, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

