Activation of endogenous retroviruses during brain development causes an inflammatory response
- PMID: 33644903
- PMCID: PMC8090857
- DOI: 10.15252/embj.2020106423
Activation of endogenous retroviruses during brain development causes an inflammatory response
Abstract
Endogenous retroviruses (ERVs) make up a large fraction of mammalian genomes and are thought to contribute to human disease, including brain disorders. In the brain, aberrant activation of ERVs is a potential trigger for an inflammatory response, but mechanistic insight into this phenomenon remains lacking. Using CRISPR/Cas9-based gene disruption of the epigenetic co-repressor protein Trim28, we found a dynamic H3K9me3-dependent regulation of ERVs in proliferating neural progenitor cells (NPCs), but not in adult neurons. In vivo deletion of Trim28 in cortical NPCs during mouse brain development resulted in viable offspring expressing high levels of ERVs in excitatory neurons in the adult brain. Neuronal ERV expression was linked to activated microglia and the presence of ERV-derived proteins in aggregate-like structures. This study demonstrates that brain development is a critical period for the silencing of ERVs and provides causal in vivo evidence demonstrating that transcriptional activation of ERV in neurons results in an inflammatory response.
Keywords: CRISPR; Trim28; brain development; microglia; transposable elements.
© 2021 The Authors. Published under the terms of the CC BY 4.0 licens.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
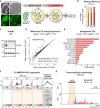
A schematic of the workflow for Trim28‐KO in mouse NPC cultures. Scale bars: embryo 1 mm, NPCs 20 μm.
Estimation of gene editing at the Trim28 loci using NGS‐sequencing of amplicons. Black bars indicate % of frameshift indels. Columns show an average of two biological replicates per guide RNA and error bars show mean ± SD.
Western blot confirmed the loss of Trim28 expression upon Trim28‐KO in mouse NPCs.
RNA‐seq analysis of the expression of TE families using TEtranscripts
The significantly upregulated TE families with a fold change larger than 0.5 upon Trim28‐KO in mouse NPCs. The dashed line indicates significance.
RNA‐seq analysis of the Trim28‐KO and control samples, visualizing full length MMERVK10C elements (left panels) and CUT&RUN analysis of H3K9me3 in mouse NPCs (right panel). The location of the full length MMERVK10Cs is indicated as a thick black line under each histogram.
Example of transcriptional readthrough outside a full length MMERVK10C into a nearby gene.

- A
Differential expression analysis of individual TEs in mouse NPCs upon CRISPR/Cas9‐mediatied disruption of Trim28 was performed with DESeq2 (described in Materials and Methods).
- B
The upregulated transcription of MMERVK10C elements was validated by qRT–PCR. Columns show an average of the individual data points.
- C
Genes located in the close vicinity of upregulated MMERVK10C elements were significantly upregulated. X‐axis indicate the window‐of‐inclusion for genes located close to an MMERVK10C. Boxplot hinges represent first and third quartile, and the median is indicated by the central band. Whiskers extend to 1.5 times the interquartile range.
- D
RNA‐seq differential expression analysis of protein coding genes performed with DESeq2 (described in Materials and Methods) in mouse NPCs upon CRISPR/Cas9‐mediatied disruption of Trim28.
- E, F
PCA analysis of gene expression was unable to distinguish the Trim28‐KO cells from Ctl, while PCA analysis of TE expression was. The spread of the groups is visualized with opaque ellipses.
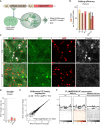
- A
A schematic of the workflow targeting Trim28 in the mouse forebrain using AAV vectors expressing the gRNA and a nuclear RFP reporter. 8 weeks later, the injected animals were analyzed either by immunohistochemical analysis or nuclei isolation by FACS prior to DNA/RNA‐sequencing.
- B
Estimation of gene editing at the Trim28 loci using NGS‐sequencing of amplicons from DNA isolated from 50,000 RFP+ nuclei per animal. One animal per group was analyzed. Black bars indicate % of the detected indels that disrupted the frameshift.
- C, D
Gene editing of the Trim28‐loci resulted in a robust loss of Trim28 protein, as evaluated by IHC where the expression of Trim28 in RFP+ cells was quantified and is displayed as mean ± SEM. Approximately 600 RFP+ cells per animal and group was evaluated. Scale bar 30 μm.
- E
RNA‐seq analysis of the expression of TE families using TEtranscripts.
- F
RNA‐seq analysis of the Trim28‐KO and control samples, visualizing full length MMERVK10C elements (left panels) and ChIP‐seq analysis of H3K9me3 in adult forebrain neurons (right panel). The location of the full length MMERVK10Cs is indicated as a thick black line under each histogram.
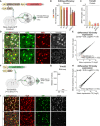
- A
A schematic of the workflow targeting Trim28 in the forebrain of Stop‐Cas9‐GFP knock‐in mice using AAV vectors expressing the gRNA and a nuclear RFP reporter as well as an AAV vector expressing Cre, specifically in neurons by the Synapsin promoter. 8 weeks after injection, the animals were analyzed either by IHC or by DNA/RNA‐sequencing following nuclei isolation by FACS.
- B
Gene editing efficiency was evaluated by amplicon sequencing of the respective targeted sequences. DNA was isolated from 50,000 RFP+ nuclei per animal, one animal per group was analyzed. Black bars indicate % of frameshift mutations.
- C, D
Neuron‐specific editing of the Trim28‐loci resulted in a robust loss of the Trim28 protein in neurons, as evaluated by IHC where the expression of Trim28 in RFP+ cells was quantified and is displayed as mean ± SEM. Approximately 550 RFP+/GFP+ cells per animal and group was evaluated. Scale bar 30 µm.
- E
Differential expression analysis of TEs in adult neurons from the Stop‐Cas9‐GFP knock‐in mice upon Trim28‐KO was done by using TEtranscripts and DESeq2 (described in Materials and Methods).
- F
A schematic of the workflow targeting Trim28 in adult neurons in the forebrain of Trim28‐flox mice (+/− and +/+) by co‐injecting AAV vectors expressing Cre or nuclear GFP under the control of the Synapsin promoter. 8 weeks after injection, the animals were analyzed either by IHC or by RNA‐sequencing following nuclei isolation by FACS.
- G
RNA‐seq analysis of the isolated GFP+ nuclei revealed a loss of the Trim28 transcript. Boxplot hinges represent first and third quartile, and the median is indicated by the central band. Whiskers extend to 1.5 times the interquartile range.
- H
TE differential expression analysis on the adult neurons in the Trim28‐flox animals was done using TEtranscripts and DESeq2 (described in Materials and Methods).
- I
IHC for Trim28, GFP, and the neuronal marker NeuN revealed a complete neuronal loss of the protein Trim28 in the Trim28‐KO animals (Trim28‐flox mice (+/+)). Scale bar 75 µm.

RNA‐seq analysis of Trim28 expression in the adult cortex of Emx1‐Cre/ Trim28‐KO animals (Emx1‐Cre(+/−), Trim28‐flox(+/+)). Boxplot hinges represent first and third quartile, and the median is indicated by the central band. Whiskers extend to 1.5 times the interquartile range.
Differential TE expression analysis for individual TEs in the adult cortex of Emx1‐Cre/Trim28‐KO animals compared to their Cre‐negative control litter mates was performed with DESeq2 (described in Materials and Methods).
Expression of genes nearby upregulated full length MMERVK10Cs were not upregulated in the Emx1‐Cre/Trim28‐KO animals. X‐axis indicate the window‐of‐inclusion for genes located close to an MMERVK10C (test performed with DESeq2, Log2FC > 0). Boxplot hinges represent first and third quartile, and the median is indicated by the central band. Whiskers extend to 1.5 times the interquartile range.
No transcriptional readthrough into nearby genes from upregulated MMERVK10Cs were observed, exemplified with an UCSC screenshot of the same location in the genome as shown in Fig 1G.
Differential gene expression performed with DESeq2 (see Materials and Methods) in the adult cortex of Emx1‐Cre/Trim28‐KO animals compared to their control litter mates.
Differential gene expression performed with DESeq2 (see Materials and Methods) in the adult forebrain upon AAV.Cre/Trim28‐KO compared to controls.
Venn diagrams showing significantly up‐ and downregulated genes in the Emx1‐Cre/Trim28‐KO and the AAV.Cre/Trim28‐KO animals, as well as the genes overlapping in between them.
GO term analysis of the overlapping upregulated genes between Emx1‐Cre/Trim28‐KO and the AAV.Cre/Trim28‐KO animals.

- A
An overview of the defined cell clusters from single cell RNA‐seq of Emx1‐Cre/Trim28‐flox animals in an UMAP plot.
- B–H
The projected expression of two cell markers per cluster on the UMAP.

The number of Iba1+ cells did not differ in the cortex of Emx1‐Cre/Trim28‐KO animals and controls, here shown as mean ± SEM, unpaired t‐test. Iba1+ cells were counted in 20 photographs (20x objective) per animal, control n = 3 and KO n = 2.
Immunohistochemical analysis for the microglia marker upon Trim28‐KO in mature neurons in vivo (AAV.Syn‐Cre/Trim28‐KO animals) revealed no difference in Iba1 morphology between Trim28‐KO animals and controls. Scale bar 75 μm.
Inflammatory genes were chosen from 216 genes annotated in the immune response GO term (GO0006954) if none of the confidence intervals overlapped zero Three genes were significantly downregulated upon the Emx1‐Cre/Trim28‐KO and are labeled in red: Bmp6, Cd163, and Ptgdr (P‐adj < 0.05), n = 3 per condition. Error lines showing 95% confidence intervals. The list of viral defense genes was retrieved from (Liu et al, 2018). None of these were significantly different upon the Emx1‐Cre/Trim28‐KO.
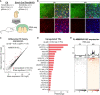
A schematic of the breeding scheme resulting in highly efficient conditional deletion of Trim28‐KO during cortical development and analyzing the adult tissue 3 months later by IHC and RNA‐seq.
IHC for Trim28 in the adult cortex revealed that the protein was lost in cells exposed to Cre‐activity during brain development (GFP+ cells). Scale bars: low magnification 75 μm, high magnification 20 µm.
RNA‐seq analysis of the expression of TE families using TEtranscripts
Significantly upregulated TE families upon the Trim28‐KO, in which the families with the highest fold change are listed.
RNA‐seq analysis of full length MMERVK10Cs in the adult tissue. The location of the full length MMERVK10Cs is indicated as a thick black line under each histogram.

- A
A schematic of the workflow for the single‐nuclei RNA‐seq of cortical tissue from Emx1‐Cre/Trim28‐KO animals and their littermates.
- B
UMAP showing the unbiased clustering analysis with seven different cell clusters.
- C, D
UMAP and pie charts showing the distribution of Trim28‐KO and control cells over the seven different clusters. There were no major differences in proportions of the different cell clusters between Emx1‐Cre/Trim28‐KO animals and controls.
- E–H
A selection of significant cell‐type‐specific changes in gene expression between Emx1‐Cre/Trim28‐KO animals and controls as revealed by single‐nuclei RNA‐seq. The black dots represent the mean value (Wilcoxon rank sum test, (P‐adj value < 0.01), n = 2. For a full list, see Table EV2.
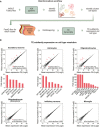
The workflow used to analyze ERV expression in single‐nuclei RNA‐seq data.
Mean plots show the changes of TE subfamily expression in each cell cluster upon the Emx1‐Cre/Trim28‐KO. A differential expression analysis performed with DESeq2 (described in material and methods) showed upregulated TEs in cell types in which Trim28 was deleted (excitatory neurons, astrocytes, oligodendrocytes, and oligodendrocyte precursors), (indicated with red dots: P‐adj < 0.01, log2FC > 3). The specific upregulated elements and their fold changes are listed in bar graphs under each mean plot. In cell types in which Trim28 was not deleted, TE expression remained unaffected (inhibitory neurons and microglia).
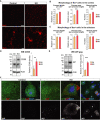
- A
IHC analysis for the microglia marker Iba1 in Emx1‐Cre/Trim28‐KO animals and controls. Scale bars: Low magnification 75 μm, high magnification 10 μm.
- B, C
The morphology of microglia in cortex and striatum (control region) of Emx1‐Cre/Trim28‐KO animals and controls were quantified by high‐content screening of Iba1 immunoreactivity, revealing differences in process length, area, and number of branch points specifically in cortex, error bars mean ± SEM (unpaired t‐test. P values: 1.8 × 10−4, 9.6 × 10−4, and 5.2 × 10−6, respectively). A large number of photographs from three control and two KO animals were analyzed, see material and methods for details.
- D
Western blot revealed an increased expression of CD68 in the Emx1‐Cre/Trim28‐KO animals (n = 5 per group), unpaired t‐test, **P = 0.0036, error bars mean ± SEM. The star indicates an unspecific band.
- E
An increased expression of the ERV‐derived protein IAP‐Gag was detected in the cortex of Emx1‐Cre/Trim28‐KO animals (n = 5 per group), unpaired t‐test, ***P = 0.0008, error bars mean ± SEM.
- F
Immunohistochemistry for IAP‐Gag visualized the presence of ERV proteins in the cortex of Emx1‐Cre/Trim28‐KO animals. The distribution of IAP‐Gag was heterogenous among Trim28‐KO neurons, where it was either accumulated in a low (#) or high (°) number of aggregate‐like structures in the cytoplasm, or as weak homogenous staining throughout the cytoplasm (#) or not present at all (*). Scale bars: low magnification 75 µm, high magnification 5 μm.
References
-
- Andrews WD, Tuke PW, Al‐Chalabi A, Gaudin P, Ijaz S, Parton MJ, Garson JA (2000) Detection of reverse transcriptase activity in the serum of patients with motor neurone disease. J Med Virol 61: 527–532 - PubMed
-
- Antony JM, van Marle G, Opii W, Butterfield DA, Mallet F, Yong VW, Wallace JL, Deacon RM, Warren K, Power C (2004) Human endogenous retrovirus glycoprotein‐mediated induction of redox reactants causes oligodendrocyte death and demyelination. Nat Neurosci 7: 1088–1095 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

