Signaling-to-chromatin pathways in the immune system
- PMID: 33644906
- PMCID: PMC8548991
- DOI: 10.1111/imr.12955
Signaling-to-chromatin pathways in the immune system
Abstract
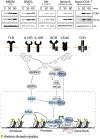
Complex organisms are able to respond to diverse environmental cues by rapidly inducing specific transcriptional programs comprising a few dozen genes among thousands. The highly complex environment within the nucleus-a crowded milieu containing large genomes tightly condensed with histone proteins in the form of chromatin-makes inducible transcription a challenge for the cell, akin to the proverbial needle in a haystack. The different signaling pathways and transcription factors involved in the transmission of information from the cell surface to the nucleus have been readily explored, but not so much the specific mechanisms employed by the cell to ultimately instruct the chromatin changes necessary for a fast and robust transcription activation. Signaling pathways rely on cascades of protein kinases that, in addition to activating transcription factors can also activate the chromatin template by phosphorylating histone proteins, what we refer to as "signaling-to-chromatin." These pathways appear to be selectively employed and especially critical for driving inducible transcription in macrophages and likely in diverse other immune cell populations. Here, we discuss signaling-to-chromatin pathways with potential relevance in diverse immune cell populations together with chromatin related mechanisms that help to "solve" the needle in a haystack challenge of robust chromatin activation and inducible transcription.
Keywords: chromatin; histone phosphorylation; immune responses; signaling.
© 2021 John Wiley & Sons A/S. Published by John Wiley & Sons Ltd.
Figures

References
-
- Kornberg RD. Chromatin structure: a repeating unit of histones and DNA. Science. 1974;184:868–871. - PubMed
-
- Van Holde K. Chromatin, New York, NY: Springer-Verlag; 1988.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

