Pseudo Natural Products-Chemical Evolution of Natural Product Structure
- PMID: 33644925
- PMCID: PMC8360037
- DOI: 10.1002/anie.202016575
Pseudo Natural Products-Chemical Evolution of Natural Product Structure
Abstract
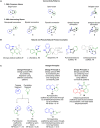
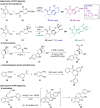
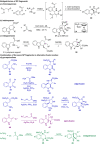
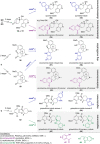
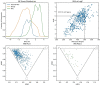
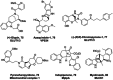
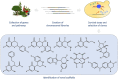
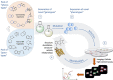
Pseudo-natural products (PNPs) combine natural product (NP) fragments in novel arrangements not accessible by current biosynthesis pathways. As such they can be regarded as non-biogenic fusions of NP-derived fragments. They inherit key biological characteristics of the guiding natural product, such as chemical and physiological properties, yet define small molecule chemotypes with unprecedented or unexpected bioactivity. We iterate the design principles underpinning PNP scaffolds and highlight their syntheses and biological investigations. We provide a cheminformatic analysis of PNP collections assessing their molecular properties and shape diversity. We propose and discuss how the iterative analysis of NP structure, design, synthesis, and biological evaluation of PNPs can be regarded as a human-driven branch of the evolution of natural products, that is, a chemical evolution of natural product structure.
Keywords: biological activity; chemical biology; fragment-based design; natural products; natural selection.
© 2021 The Authors. Angewandte Chemie International Edition published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest. G.K. is now an employee of AstraZeneca, U.K.
Figures



Similar articles
-
Chemical Evolution of Natural Product Structure.J Am Chem Soc. 2022 Mar 2;144(8):3314-3329. doi: 10.1021/jacs.1c11270. Epub 2022 Feb 21. J Am Chem Soc. 2022. PMID: 35188375 Free PMC article.
-
Guided by evolution: from biology oriented synthesis to pseudo natural products.Nat Prod Rep. 2020 Nov 18;37(11):1497-1510. doi: 10.1039/d0np00015a. Nat Prod Rep. 2020. PMID: 33020792 Review.
-
Chromopynones are pseudo natural product glucose uptake inhibitors targeting glucose transporters GLUT-1 and -3.Nat Chem. 2018 Nov;10(11):1103-1111. doi: 10.1038/s41557-018-0132-6. Epub 2018 Sep 10. Nat Chem. 2018. PMID: 30202104
-
Natural product fragment combination to performance-diverse pseudo-natural products.Nat Commun. 2021 Mar 25;12(1):1883. doi: 10.1038/s41467-021-22174-4. Nat Commun. 2021. PMID: 33767198 Free PMC article.
-
Principle and design of pseudo-natural products.Nat Chem. 2020 Mar;12(3):227-235. doi: 10.1038/s41557-019-0411-x. Epub 2020 Feb 3. Nat Chem. 2020. PMID: 32015480 Review.
Cited by
-
A divergent intermediate strategy yields biologically diverse pseudo-natural products.Nat Chem. 2024 Jun;16(6):945-958. doi: 10.1038/s41557-024-01458-4. Epub 2024 Feb 16. Nat Chem. 2024. PMID: 38365941 Free PMC article.
-
Computational Approaches to Enzyme Inhibition by Marine Natural Products in the Search for New Drugs.Mar Drugs. 2023 Jan 30;21(2):100. doi: 10.3390/md21020100. Mar Drugs. 2023. PMID: 36827141 Free PMC article. Review.
-
Identification of readily available pseudo-natural products.RSC Med Chem. 2024 Jul 4;15(8):2709-2717. doi: 10.1039/d4md00310a. eCollection 2024 Aug 14. RSC Med Chem. 2024. PMID: 39149091 Free PMC article.
-
Time-Dependent Comparison of the Structural Variations of Natural Products and Synthetic Compounds.Int J Mol Sci. 2024 Oct 25;25(21):11475. doi: 10.3390/ijms252111475. Int J Mol Sci. 2024. PMID: 39519027 Free PMC article.
-
The Time and Place for Nature in Drug Discovery.JACS Au. 2022 Oct 14;2(11):2400-2416. doi: 10.1021/jacsau.2c00415. eCollection 2022 Nov 28. JACS Au. 2022. PMID: 36465532 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

