Crystal structures of adenylylated and unadenylylated PII protein GlnK from Corynebacterium glutamicum
- PMID: 33645536
- PMCID: PMC7919409
- DOI: 10.1107/S2059798321000735
Crystal structures of adenylylated and unadenylylated PII protein GlnK from Corynebacterium glutamicum
Abstract
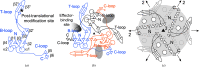
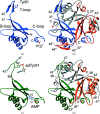
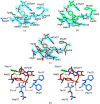
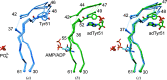
PII proteins are ubiquitous signaling proteins that are involved in the regulation of the nitrogen/carbon balance in bacteria, archaea, and some plants and algae. Signal transduction via PII proteins is modulated by effector molecules and post-translational modifications in the PII T-loop. Whereas the binding of ADP, ATP and the concomitant binding of ATP and 2-oxoglutarate (2OG) engender two distinct conformations of the T-loop that either favor or disfavor the interaction with partner proteins, the structural consequences of post-translational modifications such as phosphorylation, uridylylation and adenylylation are far less well understood. In the present study, crystal structures of the PII protein GlnK from Corynebacterium glutamicum have been determined, namely of adenylylated GlnK (adGlnK) and unmodified unadenylylated GlnK (unGlnK). AdGlnK has been proposed to act as an inducer of the transcription repressor AmtR, and the adenylylation of Tyr51 in GlnK has been proposed to be a prerequisite for this function. The structures of unGlnK and adGlnK allow the first atomic insights into the structural implications of the covalent attachment of an AMP moiety to the T-loop. The overall GlnK fold remains unaltered upon adenylylation, and T-loop adenylylation does not appear to interfere with the formation of the two major functionally important T-loop conformations, namely the extended T-loop in the canonical ADP-bound state and the compacted T-loop that is adopted upon the simultaneous binding of Mg-ATP and 2OG. Thus, the PII-typical conformational switching mechanism appears to be preserved in GlnK from C. glutamicum, while at the same time the functional repertoire becomes expanded through the accommodation of a peculiar post-translational modification.
Keywords: AMPylation; T-loop conformations; adenylylation; bacterial signal transduction; crystal structures; nitrogen starvation; post-translational modifications.
open access.
Figures
Similar articles
-
Structure of AmtR, the global nitrogen regulator of Corynebacterium glutamicum, in free and DNA-bound forms.FEBS J. 2016 Mar;283(6):1039-59. doi: 10.1111/febs.13643. Epub 2016 Feb 10. FEBS J. 2016. PMID: 26744254
-
Similarities in the structure of the transcriptional repressor AmtR in two different space groups suggest a model for the interaction with GlnK.Acta Crystallogr F Struct Biol Commun. 2017 Mar 1;73(Pt 3):146-151. doi: 10.1107/S2053230X17002485. Epub 2017 Feb 21. Acta Crystallogr F Struct Biol Commun. 2017. PMID: 28291750 Free PMC article.
-
PII Signal Transduction Protein GlnK Alleviates Feedback Inhibition of N-Acetyl-l-Glutamate Kinase by l-Arginine in Corynebacterium glutamicum.Appl Environ Microbiol. 2020 Apr 1;86(8):e00039-20. doi: 10.1128/AEM.00039-20. Print 2020 Apr 1. Appl Environ Microbiol. 2020. PMID: 32060028 Free PMC article.
-
The role of effector molecules in signal transduction by PII proteins.Biochem Soc Trans. 2011 Jan;39(1):189-94. doi: 10.1042/BST0390189. Biochem Soc Trans. 2011. PMID: 21265771 Review.
-
Nitrogen control in Corynebacterium glutamicum: proteins, mechanisms, signals.J Microbiol Biotechnol. 2007 Feb;17(2):187-94. J Microbiol Biotechnol. 2007. PMID: 18051748 Review.
Cited by
-
Transcriptomic and enzymatic analysis reveals the roles of glutamate dehydrogenase in Corynebacterium glutamicum.AMB Express. 2022 Dec 28;12(1):161. doi: 10.1186/s13568-022-01506-7. AMB Express. 2022. PMID: 36576637 Free PMC article.
References
-
- Beckers, G., Strösser, J., Hildebrandt, U., Kalinowski, J., Farwick, M., Krämer, R. & Burkovski, A. (2005). Mol. Microbiol. 58, 580–595. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

