Genome-wide interference of ZNF423 with B-lineage transcriptional circuitries in acute lymphoblastic leukemia
- PMID: 33646306
- PMCID: PMC7948270
- DOI: 10.1182/bloodadvances.2020001844
Genome-wide interference of ZNF423 with B-lineage transcriptional circuitries in acute lymphoblastic leukemia
Abstract
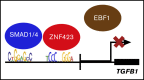
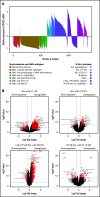
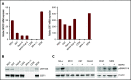
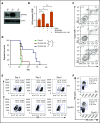
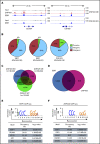
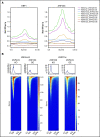
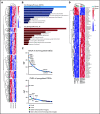
Aberrant expression of the transcriptional modulator and early B-cell factor 1 (EBF1) antagonist ZNF423 has been implicated in B-cell leukemogenesis, but its impact on transcriptional circuitries in lymphopoiesis has not been elucidated in a comprehensive manner. Herein, in silico analyses of multiple expression data sets on 1354 acute leukemia samples revealed a widespread presence of ZNF423 in various subtypes of acute lymphoblastic leukemia (ALL). Average expression of ZNF423 was highest in ETV6-RUNX1, B-other, and TCF3-PBX1 ALL followed by BCR-ABL, hyperdiploid ALL, and KMT2A-rearranged ALL. In a KMT2A-AFF1 pro-B ALL model, a CRISPR-Cas9-mediated genetic ablation of ZNF423 decreased cell viability and significantly prolonged survival of mice upon xenotransplantation. For the first time, we characterized the genome-wide binding pattern of ZNF423, its impact on the chromatin landscape, and differential gene activities in a B-lineage context. In general, chromatin-bound ZNF423 was associated with a depletion of activating histone marks. At the transcriptional level, EBF1-dependent transactivation was disrupted by ZNF423, whereas repressive and pioneering activities of EBF1 were not discernibly impeded. Unexpectedly, we identified an enrichment of ZNF423 at canonical EBF1-binding sites also in the absence of EBF1, which was indicative of intrinsic EBF1-independent ZNF423 activities. A genome-wide motif search at EBF1 target gene loci revealed that EBF1 and ZNF423 co-regulated genes often contain SMAD1/SMAD4-binding motifs as exemplified by the TGFB1 promoter, which was repressed by ZNF423 outcompeting EBF1 by depending on its ability to bind EBF1 consensus sites and to interact with EBF1 or SMADs. Overall, these findings underscore the wide scope of ZNF423 activities that interfere with B-cell lymphopoiesis and contribute to leukemogenesis.
© 2021 by The American Society of Hematology.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Figures

References
-
- Kuiper RP, Schoenmakers EF, van Reijmersdal SV, et al. High-resolution genomic profiling of childhood ALL reveals novel recurrent genetic lesions affecting pathways involved in lymphocyte differentiation and cell cycle progression. Leukemia. 2007;21(6):1258-1266. - PubMed
-
- Mullighan CG, Goorha S, Radtke I, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446(7137):758-764. - PubMed
-
- Mandel EM, Grosschedl R. Transcription control of early B cell differentiation. Curr Opin Immunol. 2010;22(2):161-167. - PubMed
-
- Parra M. Epigenetic events during B lymphocyte development. Epigenetics. 2009;4(7):462-468. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

