DNA methylation meta-analysis reveals cellular alterations in psychosis and markers of treatment-resistant schizophrenia
- PMID: 33646943
- PMCID: PMC8009672
- DOI: 10.7554/eLife.58430
DNA methylation meta-analysis reveals cellular alterations in psychosis and markers of treatment-resistant schizophrenia
Abstract
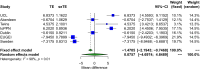
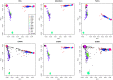
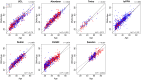
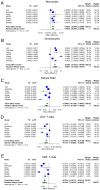
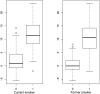
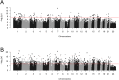
We performed a systematic analysis of blood DNA methylation profiles from 4483 participants from seven independent cohorts identifying differentially methylated positions (DMPs) associated with psychosis, schizophrenia, and treatment-resistant schizophrenia. Psychosis cases were characterized by significant differences in measures of blood cell proportions and elevated smoking exposure derived from the DNA methylation data, with the largest differences seen in treatment-resistant schizophrenia patients. We implemented a stringent pipeline to meta-analyze epigenome-wide association study (EWAS) results across datasets, identifying 95 DMPs associated with psychosis and 1048 DMPs associated with schizophrenia, with evidence of colocalization to regions nominated by genetic association studies of disease. Many schizophrenia-associated DNA methylation differences were only present in patients with treatment-resistant schizophrenia, potentially reflecting exposure to the atypical antipsychotic clozapine. Our results highlight how DNA methylation data can be leveraged to identify physiological (e.g., differential cell counts) and environmental (e.g., smoking) factors associated with psychosis and molecular biomarkers of treatment-resistant schizophrenia.
Keywords: DNA methylation; clozapine; epigenetics; genetics; genomics; human; psychosis; schizophrenia.
© 2021, Hannon et al.
Conflict of interest statement
EH, ED, GM, JB, NB, MB, AC, CC, DD, TD, GD, MG, AG, CG, HH, CH, VJ, RK, JK, GK, CM, AM, DM, KM, CM, IN, DQ, AR, BR, DS, ST, TT, JV, JW, EV, GB, LS, JM No competing interests declared, MD M Di Forti reports personal fees from Janssen, outside the submitted work. FG Fiona Gaughran has received honoraria from Lundbeck, Otsuka, and Sunovion. She has a family member with professional links to Lilly and GSK, including shares. KK Kaarina Kowalec has consulted with Emerald Lake Safety Ltd. (2017-2018) and has received speaker honoraria from Biogen/Fraser Health Multiple Sclerosis Clinic (2018). Both are unrelated to the work published here. JM James MacCabe has received research funding from H Lundbeck. MO Michael C O'Donovan is supported by a collaborative research grant from Takeda Pharmaceuticals. Takeda played no part in the conception, design, implementation, or interpretation of this study. PS PF Sullivan reports the following potentially competing financial interests. Current: Lundbeck (advisory committee, grant recipient). Past three years: Pfizer (scientific advisory board). DC David A Collier is a full time employee and stockholder of Eli Lilly and Company. RM Robin M Murray reports personal fees from Janssen, Lundbeck, Sunovion, Recordati and Otsuka
Figures














References
-
- Aberg KA, McClay JL, Nerella S, Clark S, Kumar G, Chen W, Khachane AN, Xie L, Hudson A, Gao G, Harada A, Hultman CM, Sullivan PF, Magnusson PK, van den Oord EJ. Methylome-wide association study of schizophrenia: identifying blood biomarker signatures of environmental insults. JAMA Psychiatry. 2014;71:255–264. doi: 10.1001/jamapsychiatry.2013.3730. - DOI - PMC - PubMed
-
- Bachmann CJ, Aagaard L, Bernardo M, Brandt L, Cartabia M, Clavenna A, Coma Fusté A, Furu K, Garuoliené K, Hoffmann F, Hollingworth S, Huybrechts KF, Kalverdijk LJ, Kawakami K, Kieler H, Kinoshita T, López SC, Machado-Alba JE, Machado-Duque ME, Mahesri M, Nishtala PS, Piovani D, Reutfors J, Saastamoinen LK, Sato I, Schuiling-Veninga CCM, Shyu YC, Siskind D, Skurtveit S, Verdoux H, Wang LJ, Zara Yahni C, Zoëga H, Taylor D. International trends in clozapine use: a study in 17 countries. Acta Psychiatrica Scandinavica. 2017;136:37–51. doi: 10.1111/acps.12742. - DOI - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
Grants and funding
- G0801418/MRC_/Medical Research Council/United Kingdom
- R01 MH077139/MH/NIMH NIH HHS/United States
- MR/K013807/1/MRC_/Medical Research Council/United Kingdom
- 085475/B/08/Z/WT_/Wellcome Trust/United Kingdom
- MR/R005176/1/MRC_/Medical Research Council/United Kingdom
- MR/M008436/1/MRC_/Medical Research Council/United Kingdom
- MR/M008924/1/MRC_/Medical Research Council/United Kingdom
- MR/L011794/1/MRC_/Medical Research Council/United Kingdom
- 090532/Z/09/Z/WT_/Wellcome Trust/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- MR/L010305/1/MRC_/Medical Research Council/United Kingdom
- G0800509/MRC_/Medical Research Council/United Kingdom
- R01 MH052857/MH/NIMH NIH HHS/United States
- 075491/Z/04/B/WT_/Wellcome Trust/United Kingdom
- RP-PG-0606-1049/DH_/Department of Health/United Kingdom
- 072894/Z/03/Z/WT_/Wellcome Trust/United Kingdom
- MR/T007818/1/MRC_/Medical Research Council/United Kingdom
- Fellowship (MR/M008436/1)/MRC_/Medical Research Council/United Kingdom
- 085475/Z/08/Z/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

