The allelic rice immune receptor Pikh confers extended resistance to strains of the blast fungus through a single polymorphism in the effector binding interface
- PMID: 33647072
- PMCID: PMC7951977
- DOI: 10.1371/journal.ppat.1009368
The allelic rice immune receptor Pikh confers extended resistance to strains of the blast fungus through a single polymorphism in the effector binding interface
Abstract
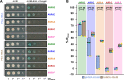
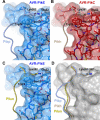
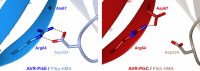
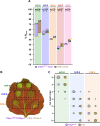
Arms race co-evolution drives rapid adaptive changes in pathogens and in the immune systems of their hosts. Plant intracellular NLR immune receptors detect effectors delivered by pathogens to promote susceptibility, activating an immune response that halts colonization. As a consequence, pathogen effectors evolve to escape immune recognition and are highly variable. In turn, NLR receptors are one of the most diverse protein families in plants, and this variability underpins differential recognition of effector variants. The molecular mechanisms underlying natural variation in effector recognition by NLRs are starting to be elucidated. The rice NLR pair Pik-1/Pik-2 recognizes AVR-Pik effectors from the blast fungus Magnaporthe oryzae, triggering immune responses that limit rice blast infection. Allelic variation in a heavy metal associated (HMA) domain integrated in the receptor Pik-1 confers differential binding to AVR-Pik variants, determining resistance specificity. Previous mechanistic studies uncovered how a Pik allele, Pikm, has extended recognition to effector variants through a specialized HMA/AVR-Pik binding interface. Here, we reveal the mechanistic basis of extended recognition specificity conferred by another Pik allele, Pikh. A single residue in Pikh-HMA increases binding to AVR-Pik variants, leading to an extended effector response in planta. The crystal structure of Pikh-HMA in complex with an AVR-Pik variant confirmed that Pikh and Pikm use a similar molecular mechanism to extend their pathogen recognition profile. This study shows how different NLR receptor alleles functionally converge to extend recognition specificity to pathogen effectors.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
- BB/M011216/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/M011216/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/M02198X/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/P012574/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

