Identification of tumor antigens and immune subtypes of pancreatic adenocarcinoma for mRNA vaccine development
- PMID: 33648511
- PMCID: PMC7917175
- DOI: 10.1186/s12943-021-01310-0
Identification of tumor antigens and immune subtypes of pancreatic adenocarcinoma for mRNA vaccine development
Abstract
Background: Although mRNA vaccines have been effective against multiple cancers, their efficacy against pancreatic adenocarcinoma (PAAD) remains undefined. Accumulating evidence suggests that immunotyping can indicate the comprehensive immune status in tumors and their immune microenvironment, which is closely associated with therapeutic response and vaccination potential. The aim of this study was to identify potent antigens in PAAD for mRNA vaccine development, and further distinguish immune subtypes of PAAD to construct an immune landscape for selecting suitable patients for vaccination.
Methods: Gene expression profiles and clinical information of 239 PAAD datasets were extracted from ICGC, and RNA-Seq data of 103 samples were retrieved from TCGA. GEPIA was used to calculate differential expression levels and prognostic indices, cBioPortal program was used to compare genetic alterations, and TIMER was used to explore correlation between genes and immune infiltrating cells. Consensus cluster was used for consistency matrix construction and data clustering, DAVID was used for functional annotation, and graph learning-based dimensional reduction was used to depict immune landscape.
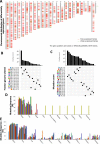
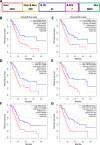
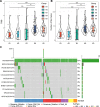
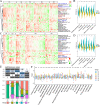
Results: Six overexpressed and mutated tumor antigens associated with poor prognosis and infiltration of antigen presenting cells were identified in PAAD, including ADAM9, EFNB2, MET, TMOD3, TPX2, and WNT7A. Furthermore, five immune subtypes (IS1-IS5) and nine immune gene modules of PAAD were identified that were consistent in both patient cohorts. The immune subtypes showed distinct molecular, cellular and clinical characteristics. IS1 and IS2 exhibited immune-activated phenotypes and correlated to better survival compared to the other subtypes. IS4 and IS5 tumors were immunologically cold and associated with higher tumor mutation burden. Immunogenic cell death modulators, immune checkpoints, and CA125 and CA199, were also differentially expressed among the five immune subtypes. Finally, the immune landscape of PAAD showed a high degree of heterogeneity between individual patients.
Conclusions: ADAM9, EFNB2, MET, TMOD3, TPX2, and WNT7A are potent antigens for developing anti-PAAD mRNA vaccine, and patients with IS4 and IS5 tumors are suitable for vaccination.
Keywords: Immune landscape; Immunotype; Pancreatic adenocarcinoma; Tumor immune microenvironment; mRNA vaccine.
Conflict of interest statement
The authors declared no potential conflicts of interest in terms of the research, authorship, and/or publication of this article.
Figures







Similar articles
-
Identification of Tumor Antigens and Immune Subtypes of Glioblastoma for mRNA Vaccine Development.Front Immunol. 2022 Feb 2;13:773264. doi: 10.3389/fimmu.2022.773264. eCollection 2022. Front Immunol. 2022. PMID: 35185876 Free PMC article.
-
Recognition of Tumor-Associated Antigens and Immune Subtypes in Glioma for mRNA Vaccine Development.Front Immunol. 2021 Sep 17;12:738435. doi: 10.3389/fimmu.2021.738435. eCollection 2021. Front Immunol. 2021. PMID: 34603319 Free PMC article.
-
Bioinformatics analyses for the identification of tumor antigens and immune subtypes of gastric adenocarcinoma.Front Genet. 2022 Dec 12;13:1068112. doi: 10.3389/fgene.2022.1068112. eCollection 2022. Front Genet. 2022. PMID: 36579327 Free PMC article.
-
Identify potential prognostic indicators and tumor-infiltrating immune cells in pancreatic adenocarcinoma.Biosci Rep. 2022 Feb 25;42(2):BSR20212523. doi: 10.1042/BSR20212523. Biosci Rep. 2022. PMID: 35083488 Free PMC article. Review.
-
Developing Vaccines in Pancreatic Adenocarcinoma: Trials and Tribulations.Curr Oncol. 2024 Aug 23;31(9):4855-4884. doi: 10.3390/curroncol31090361. Curr Oncol. 2024. PMID: 39329989 Free PMC article. Review.
Cited by
-
Sex-related disparities in outcomes of cholangiocarcinoma patients in treatment trials.Front Oncol. 2022 Aug 11;12:963753. doi: 10.3389/fonc.2022.963753. eCollection 2022. Front Oncol. 2022. PMID: 36033540 Free PMC article. No abstract available.
-
Unveiling the molecular features, relevant immune and clinical characteristics of SIGLEC15 in thyroid cancer.Front Immunol. 2022 Sep 9;13:975787. doi: 10.3389/fimmu.2022.975787. eCollection 2022. Front Immunol. 2022. PMID: 36159823 Free PMC article.
-
Identification of tumor antigens and immune subtypes in head and neck squamous cell carcinoma for mRNA vaccine development.Front Cell Dev Biol. 2022 Nov 17;10:1064754. doi: 10.3389/fcell.2022.1064754. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36467412 Free PMC article.
-
Identification of pancreatic adenocarcinoma immune subtype associated with tumor neoantigen from aberrant alternative splicing.J Gastrointest Oncol. 2024 Jun 30;15(3):1179-1197. doi: 10.21037/jgo-24-340. Epub 2024 Jun 27. J Gastrointest Oncol. 2024. PMID: 38989416 Free PMC article.
-
Tumor immune microenvironment-based therapies in pancreatic ductal adenocarcinoma: time to update the concept.J Exp Clin Cancer Res. 2024 Jan 2;43(1):8. doi: 10.1186/s13046-023-02935-3. J Exp Clin Cancer Res. 2024. PMID: 38167055 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

