Single-cell RNA sequencing in cancer research
- PMID: 33648534
- PMCID: PMC7919320
- DOI: 10.1186/s13046-021-01874-1
Single-cell RNA sequencing in cancer research
Abstract
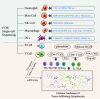
Single-cell RNA sequencing (scRNA-seq), a technology that analyzes transcriptomes of complex tissues at single-cell levels, can identify differential gene expression and epigenetic factors caused by mutations in unicellular genomes, as well as new cell-specific markers and cell types. scRNA-seq plays an important role in various aspects of tumor research. It reveals the heterogeneity of tumor cells and monitors the progress of tumor development, thereby preventing further cellular deterioration. Furthermore, the transcriptome analysis of immune cells in tumor tissue can be used to classify immune cells, their immune escape mechanisms and drug resistance mechanisms, and to develop effective clinical targeted therapies combined with immunotherapy. Moreover, this method enables the study of intercellular communication and the interaction of tumor cells and non-malignant cells to reveal their role in carcinogenesis. scRNA-seq provides new technical means for further development of tumor research and is expected to make significant breakthroughs in this field. This review focuses on the principles of scRNA-seq, with an emphasis on the application of scRNA-seq in tumor heterogeneity, pathogenesis, and treatment.
Keywords: Immune escape; Invasion and metastasis; Single‐cell RNA sequencing; Tumor heterogeneity; Tumor microenvironment.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Wu C, Li M, Meng H, Liu Y, Niu W, Zhou Y, et al. Analysis of status and countermeasures of cancer incidence and mortality in China. Sci China Life Sci. 2019;62:640–47. 10.1007/s11427-018-9461-5. - PubMed
-
- Xiao L, Wei F, Liang F, Li Q, Deng H, Tan S, et al. TSC22D2 identified as a candidate susceptibility gene of multi-cancer pedigree using genome-wide linkage analysis and whole-exome sequencing. Carcinogenesis. 2019;40:819–27. 10.1093/carcin/bgz095. - PubMed
Publication types
MeSH terms
Grants and funding
- 81903138/National Natural Science Foundation of China
- 81972776/National Natural Science Foundation of China
- 81803025/National Natural Science Foundation of China
- 81772928/National Natural Science Foundation of China
- 81702907/National Natural Science Foundation of China
- 81772901/National Natural Science Foundation of China
- 81672993/National Natural Science Foundation of China
- 81672683/National Natural Science Foundation of China
- 2019JJ50778/Natural Science Foundation of Hunan Province
- 2018SK21210/Natural Science Foundation of Hunan Province
- 2018SK21211/Natural Science Foundation of Hunan Province
- 2018JJ3704/Natural Science Foundation of Hunan Province
- 2018JJ3815/Natural Science Foundation of Hunan Province
- 2020zzts771/Fundamental Research Funds for Central Universities of the Central South University
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

